Review of Immunology
Roger W. Melvold
Although immunology is a relative newcomer among the sciences, its phenomena have long been recognized and manipulated. Ancient peoples understood that survivors of particular diseases were protected from those diseases for the remainder of their lives, and the ancient Chinese and Egyptians even practiced forms of immunization. Surgeons have also long understood that tissues and organs would not survive when exchanged between different individuals (e.g., from cadaver donors) but could succeed when transplanted from one site to another within the same individual. However, only during the past century have the mechanisms of the immune system been illuminated, at least in part. Keep in mind that the immune system, as we usually think of it, comprises the body’s second and third lines of defense. The first line of defense consists of a number of physical barriers, including the skin and mucous membranes, the fatty acids of the skin, the low pH of the stomach, enzymes and microcidal molecules secreted by a variety of cell types, resident microbial populations, and cells that act nonspecifically against infectious organisms (1).
The Innate Immune System
Pathogen-Associated Molecular Patterns, Pattern Recognition Receptors, and Stress Molecules
When microbes breach these barriers, they are confronted with the second line of defense, the innate immune system, and, if necessary, the third line, the adaptive immune system. The innate immune system is the first of these two systems to react (2). By relying on preformed receptors, the innate immune system is able to take immediate action on contact with microbial invaders. It is only in recent years that the innate immune system’s vital role and its operational mechanisms have become appreciated (3). The innate system utilizes a set of receptors that are “hard-wired” into our genomes as a result of evolution and selection over the long period of divergence between the microbial world and our own. These receptors, called pattern recognition receptors (PRRs), are not diversified somatically within each individual by chromosomal rearrangement or junctional diversity, as are the antigen receptors of T and B
lymphocytes. PRRs recognize and bind two types of molecules: (a) molecules that are synthesized (often widely expressed) by microbes, but not by host cells, and (b) “stress signals” produced by infected or injured host cells. The first of these are called pathogen-associated molecular patterns (PAMPs) that are characteristically displayed on a broad range of microbial cells. Prominent examples among the extensive array of PAMPs include lipopolysaccharide (LPS), peptidoglycan, and flagellin. Stress signals, on the other hand, result from the expression of host cell genes in response to stress induced by insults such as infection, injury, and neoplastic transformation. In humans, stress signals include the MHC class I chain-related protein A (MICA) and MICB molecules (4).
lymphocytes. PRRs recognize and bind two types of molecules: (a) molecules that are synthesized (often widely expressed) by microbes, but not by host cells, and (b) “stress signals” produced by infected or injured host cells. The first of these are called pathogen-associated molecular patterns (PAMPs) that are characteristically displayed on a broad range of microbial cells. Prominent examples among the extensive array of PAMPs include lipopolysaccharide (LPS), peptidoglycan, and flagellin. Stress signals, on the other hand, result from the expression of host cell genes in response to stress induced by insults such as infection, injury, and neoplastic transformation. In humans, stress signals include the MHC class I chain-related protein A (MICA) and MICB molecules (4).
PRRs can be found on external cell membranes, intracellular membranes, and even in soluble form (as in the case of certain complement components). In mammals, some PRRs belong to the group of receptors known as the Toll–like receptors (TLR) that are found predominantly on phagocytic cells (e.g., dendritic cells, macrophages, neutrophils) and on some other cell types including certain endothelial and epithelial cells (5). Numerous different TLRs have been identified thus far in humans (Table 1.1), some of them located on the cell membrane and capable of detecting stimuli arriving from outside the cell, and others located on endosomal membranes within the cell and capable of detecting PAMPs derived from microbes or microbial products that are already within the cell. Some PRRs are found in soluble form, such as certain components of the complement system. All normal humans express the same array of TLRs (6,7).
Table 1.1 Mammalian (Human and/or Mouse) Toll-like Receptors (TLR) | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Phagocytes
Binding of PAMPs to membrane-bound PRRs causes activation of those cells. Activated phagocytes enlarge, increase their production of particular cytokines (e.g., IL-1, -6, -8, -12, and TNF-α), increase their production of antimicrobial molecules, increase the rate at which they ingest and degrade microbes, and begin to actively engage in hunting down additional microbes (3,4,8). The ingested and degraded material may be recycled to the cell surface by a limited subset of phagocytes (e.g., dendritic cells and macrophages) known as
antigen-presenting cells (APCs) (9). When “presented” appropriately on the surfaces of these cells, the degraded material can be detected and bound by the antigenic receptors on T lymphocytes. This binding, together with additional signals exchanged between the APCs and the T lymphocytes, can lead to activation of the T lymphocytes and the initiation of adaptive immune responses, a process described later in this chapter.
antigen-presenting cells (APCs) (9). When “presented” appropriately on the surfaces of these cells, the degraded material can be detected and bound by the antigenic receptors on T lymphocytes. This binding, together with additional signals exchanged between the APCs and the T lymphocytes, can lead to activation of the T lymphocytes and the initiation of adaptive immune responses, a process described later in this chapter.
Natural Killer Cells
Natural killer (NK) cells appear to distinguish between normal cells and those altered by neoplastic transformation or infection by some viruses, and to preferentially kill the altered cells (10,11). They do not express the types of specific antigen receptors seen on T and B lymphocytes, but instead make this distinction by detecting the presence of stress molecules. Their activity is heightened by the presence of cytokines IFN-γ and IL-2. Using receptors called killer activation receptors (KAR), NK cells recognize and bind to infected or transformed cells expressing the MICA or MICB stress molecules. Using another set of receptors called killer inhibition receptors (KIR), they then assess the expression of class I MHC molecules on the surface of those cells. MHC class I expression can be reduced or absent on transformed cells or on those infected by certain types of viruses. If MHC I expression is subnormal, killing of the targeted cells proceeds. If, however, the MHC I levels are normal, the NK cells terminate their killing program and release the targeted cells.
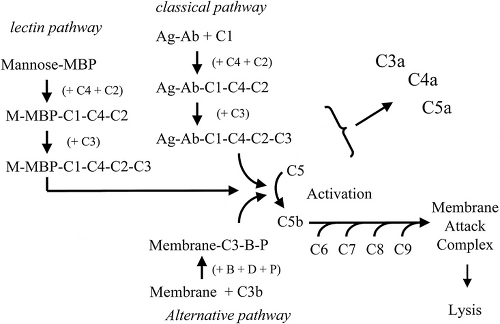
The Mannose Binding Lectin and Alternative Pathways of Complement Activation
Complement is the composite term for a number of serum proteins (complement components) that can interact with one another, as well as with antibodies under some circumstances, to produce several different chemical signals and destructive responses (3,4,12). There are three pathways for activation (Fig. 1.1). The complement components act on one another sequentially (the complement “cascade”). The cascade begins with the binding of either component C1 to an antigen–antibody complex or of component C3 to a bacterial or other membrane surface (without the assistance of antibody). The classical pathway is initiated by the binding of certain isotypes and subclasses of antibodies (IgM, IgG1, IgG2, IgG3 ) to antigen and then to complement component C1. Because of the involvement of antibodies, this pathway is usually not considered part of the innate immune system and will instead be discussed in more detail later. However, the alternative pathway and the MBL (mannose-binding lectin) pathway (also called the lectin pathway) begin with the binding of PAMPs on microbial surfaces by two molecules, C3 and mannose-binding protein (MBP), that serve as soluble PRRs. The direct binding of C3 (actually, by the C3b fragment of C3) to a microbial surface initiates the alternative pathway (involving the additional binding of components D, B, and P). The alternative pathway generates both C3 convertase that cleaves C3 itself into C3a and C3b, and serves as an amplification pathway for the generation of large amounts of fragment C3b. C3b bound to a microbial surface also serves as a potent opsonin, and phagocytic destruction of C3b-bound microbes is heightened. A third pathway for complement activation, the MBL pathway/lectin pathway, begins with the binding of MBP to mannose on bacterial cell surfaces. This pathway also leads to formation of a C3 convertase that can cleave C3 into C3a and C3b. Both the alternative and MBL pathways also result in the generation of a C5 convertase that cleaves and activates component C5 into fragments C5a and C5b. C5b can go on to bind to initiate another cascade that results in the binding of components C5b, C6, C7, C8, and C9 to a cell surface. The completion of this combination (C5b through C9) is termed the membrane attack complex and results in the rupture of the cell surface to which it is attached (3,4,12).
As complement components interact with one another, each is cleaved into fragments. Some become enzymatically active to continue the cascade. The smaller fragments (anaphylotoxins) gain hormone-like functions and are important in stimulating various inflammatory reactions. C5a (a fragment of C5) attracts neutrophils and macrophages to the site of interest. C3a (a fragment of C3) increases vascular permeability and stimulates basophils, mast cells, and platelets to release histamine and other chemicals contributing to inflammation. C4a (fragment of C4) has activity similar to C5a, although less effective. C3b (the larger fragment of C3) stimulates the phagocytic uptake of the C3b-bound microbes.
Like the nervous and endocrine systems, the immune system is adaptive, specific, and communicative. It recognizes and responds to changes in the environment, and it displays memory by adapting or altering its response to previously encountered stimuli. It can detect the presence of millions of different substances (antigens) and has an exquisite ability to discriminate among closely related molecules. Communication and interaction, involving both direct contact and soluble mediators, must occur among a variety of lymphoid and other cells for optimal function.
The complexity of the immune system is extended by genetic differences among individuals. This is because the “repertoire” of immune responses varies among unrelated individuals in an outbred, genetically heterogeneous species such as our own. Furthermore, each of us, in a sense, is “immunologically incomplete” because none of us is able to recognize and respond to all of the possible antigens that exist. Several factors contribute to this: (a) genetic or environmentally induced conditions that nonspecifically diminish immune functions, (b) variation among individuals in the genes encoding the antigen receptors of lymphocytes, (c) genetically encoded differences among individuals (often determined by the highly polymorphic genes of the human leukocyte antigen [HLA] complex) that influence whether and how the individual will respond to specific antigens, and (d) the fact that each individual’s immune system must differentiate between self (those substances that are a normal part of the body) and foreign, or nonself, to avoid autoimmunity. However, because self differs from one individual to the next, what is foreign also differs among individuals.
Antigens
Antigens were initially defined as substances identified and bound by antibodies (immunoglobulins) produced by B lymphocytes. However, because the specific antigen receptors of T lymphocytes are not immunoglobulins, the definition must be broadened to include substances that can be specifically recognized by the receptors of T or B lymphocytes or both. It is estimated that the immune system can specifically recognize at least 106 to 107 different antigens. These include both substances that are foreign to the body (nonself) and substances that are normal constituents of the body (self).
The immune system must distinguish between nonself and self antigens so that, under normal conditions, it will attack the former but not the latter. Thus, the immune system should be tolerant of self but intolerant of nonself. Autoimmune diseases arise when such distinctions are lost and the immune system attacks self antigens, a phenomenon originally described by Paul Erlich as horror autotoxicus. Well-known examples include rheumatoid arthritis, psoriasis, systemic lupus erythematosus, and some forms of diabetes.
Antigens can be divided into three general types—immunogens, haptens, and tolerogens—depending on the way in which they stimulate and interact with the immune system (3,4,13). An immunogen can, by itself, both stimulate an immune response and subsequently serve as a target of that response. The terms immunogen and antigen are often, but inappropriately, used interchangeably. A hapten cannot, by itself, stimulate an immune response. However, if a hapten is attached to a larger immunogenic molecule (a “carrier”), responses can be stimulated against both the carrier and the hapten, and the hapten itself can subsequently serve as the target of a response so provoked. A tolerogen is a substance that, after an initial exposure to the immune system, inhibits future responses against itself.
Because of the genetic diversity among individuals, a substance that is an immunogen for one person may be a tolerogen for another and may be ignored completely by the immune system of yet another. Also, a substance that acts as an immunogen when administered by one route (e.g., intramuscularly) may act as a tolerogen when applied by a different route (e.g., intragastrically), in a different form (e.g., denatured), or following treatment of the individual with therapeutic agents.
Antigens are usually protein or carbohydrate in nature and may be found as free single molecules or as parts of larger structures (e.g., expressed on the surface of an infectious agent). Although some antigens are very small and simple, others are large and complex, containing many different sites that can be individually identified by lymphocyte receptors or free immunoglobulins. Each such individual part of an antigen that can be distinctly identified by the immune system is called an epitope or determinant (i.e., the smallest identifiable antigenic unit). Thus, a single large antigen may contain many different epitopes. In general, the more complex the molecule and the greater the number of epitopes it displays, the more potent it is as an immunogen.
Adjuvants are substances that, when administered together with an immunogen (or a hapten coupled to an immunogen), enhance the response against it. For example, immunogens may be suspended in mixtures (e.g., colloidal suspensions of mycobacterial proteins
and oil) that induce localized inflammation and aid in arousal of the immune system.
and oil) that induce localized inflammation and aid in arousal of the immune system.
Molecules of the Adaptive Immune System
Immunoglobulin
B lymphocytes synthesize receptors (immunoglobulins) able to recognize and bind specific structures (antigens, determinants, epitopes). All immunoglobulins produced by a single B cell, or by a clonally derived set of B cells, have the same specificity and are able to recognize and bind only a single antigen or epitope (3,4). Immunoglobulin exists either as a surface membrane-bound molecule or in a secreted form by B cells that have been appropriately stimulated and matured.
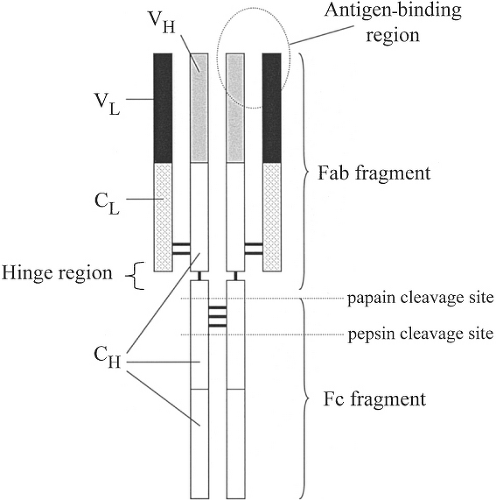
The immunoglobulin molecule is a glycoprotein composed of two identical light chains and two identical heavy chains (Fig. 1.2) linked by disulfide bonds (3,4). Enzymatic cleavage of the immunoglobulin molecule creates defined fragments. Papain produces two antigen-binding fragments (Fab) and one crystallizable fragment (Fc). Pepsin produces only a divalent antigen-binding fragment termed F(ab′)2, and the remainder of the molecule tends to be degraded and lost.
Each chain (heavy and light) contains one or more constant regions (CH or CL) and a variable region (VH or VL). Together, the variable regions of the light and heavy chains contribute to the antigen-binding sites (Fab) of the immunoglobulin molecule. The constant regions of the heavy chain (particularly in the Fc portion) determine what subsequent interactions may occur between the bound immunoglobulin and other cells or molecules of the immune system. When the antigen-binding sites are filled, a signal is transmitted through the immunoglobulin molecule, which results in conformational changes in the Fc portion of the heavy chain. These conformational changes permit the Fc portion to then interact with other molecules and cells. The conformationally altered Fc may be recognized by receptors (Fc receptors [FcR]) on macrophages and other cells, which allow them to distinguish bound from unbound immunoglobulin molecules (3,4), increasing their efficiency of phagocytosis. Other conformational changes in the Fc portion of bound immunoglobulin permit the binding of complement component C1q to initiate the classic pathway of complement activation. The Fab and F(ab′)2 fragments are useful experimental and therapeutic tools that can bind antigens without the ensuing consequences resulting from the presence of the Fc region.
Immunoglobulin light chains contain one of two types of constant regions, κ or λ. The constant regions of the heavy chains exist in five major forms (Table 1.2), each associated with a particular immunoglobulin isotype or class: Cα (immunoglobulin A [IgA]), Cδ (IgD), Cε (IgE), Cγ (IgG), and Cμ (IgM). Some of these can be subdivided into subclasses (e.g., IgG1, IgG2, IgG3, and IgG4 in humans). In the most relevant experimental animal model, the mouse, the Ig subtypes are IgG1, IgG2a, IgG2b, IgG3; however, similarly numbered subtypes in the two species are not necessarily comparable in function. Each normal individual can generate all of the isotypes. Within a single immunoglobulin molecule, both light chains are identical and of the same type (both κ or both λ), and the two heavy chains are likewise identical and of the same isotype. IgD, IgG, and IgE exist only as monomeric basic immunoglobulin units (two heavy chains and two light chains), but serum IgM exists as a pentamer of five basic units united by a J (joining) chain. IgA can be found in a variety of forms (monomers, dimers, trimers, tetramers), but is most commonly seen as a monomer (in serum) or as a dimer (in external body fluids, such as mucus, tears, and saliva). The dimeric form contains two basic units bound together by a J chain. In passing through specialized epithelial cells to external fluids, it also adds a “secretory piece,” which increases its resistance to degradation by external enzymes (3,4,14).
Table 1.2 Immunoglobulin Isotypes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
In addition to antigen-binding specificity, variability among immunoglobulin molecules derives from three other sources: allotypes, isotypes, and idiotypes. Allotypes are dictated by minor amino acid sequence differences in the constant regions of heavy or light chains, which result from slight polymorphisms in the genes encoding these molecules. Allotypic differences typically do not affect the function of the molecule and segregate within families like typical mendelian traits. Isotypes, as already discussed, are determined by more substantial differences in the heavy chain constant regions affecting the functional properties of the immunoglobulins (Table 1.2). Finally, many antigenic
determinants may be bound in more than one way, and thus there may be multiple, structurally distinct immunoglobulins with the same antigenic specificity. These differences within the antigen-binding domains of immunoglobulins that bind the same antigenic determinants are termed idiotypes.
determinants may be bound in more than one way, and thus there may be multiple, structurally distinct immunoglobulins with the same antigenic specificity. These differences within the antigen-binding domains of immunoglobulins that bind the same antigenic determinants are termed idiotypes.
Generation of Antigen-Binding Diversity among Immunoglobulins
Each immunoglobulin chain, light and heavy, is encoded not by a single gene but by a series of genes occurring in clusters on chromosomes 2, 14, and 22 (3,4,15). In humans, the series of genes encoding κ light chains, the series encoding λ light chains, and the series encoding heavy chains are all located on separate chromosomes. Within each series, the genes are found in clusters, each containing a set of similar, but not identical, genes. All of the genes are present in embryonic and germ cells and in cells other than B lymphocytes. When a cell becomes committed to the B-lymphocyte lineage, it rearranges the DNA encoding its light and heavy chains (3,4,12,16) by clipping out and degrading some of the DNA sequences. Each
differentiating B cell chooses either the κ series or the λ series (but not both). In addition, although both the maternally and paternally derived chromosomes carry these sets of genes, each B cell uses only one of them (either paternal or maternal) to produce a functional chain, a phenomenon termed allelic exclusion.
differentiating B cell chooses either the κ series or the λ series (but not both). In addition, although both the maternally and paternally derived chromosomes carry these sets of genes, each B cell uses only one of them (either paternal or maternal) to produce a functional chain, a phenomenon termed allelic exclusion.
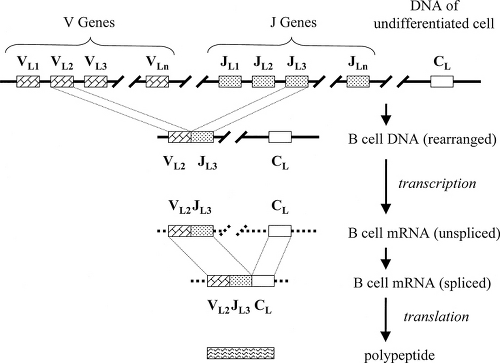
For the light chains, there are three distinct clusters of genes that contribute to the synthesis of the entire polypeptide: variable genes (VL), joining genes (JL), and constant genes (CL) (Fig. 1.3). In addition, each V gene is preceded by a leader sequence encoding a portion of the polypeptide that is important during the synthetic process but is removed when the molecule becomes functional. The VL and DL genes are used to produce the variable domain of the light chain. This is accomplished by the random selection of a single VL gene and a single JL gene to be united (VL–JL) by splicing out and discarding the intervening DNA. Henceforth, that cell and all of its clonal descendants are committed to that particular VL–JL combination. Messenger RNA for the light chain is transcribed to include the VL–JL genes, the CL gene or genes, and the intervening DNA between them. Before translation, the messenger RNA (mRNA) is spliced to unite the VL–JL genes with a CL gene so that a single continuous polypeptide can be produced from three genes that were originally separated on the chromosome.
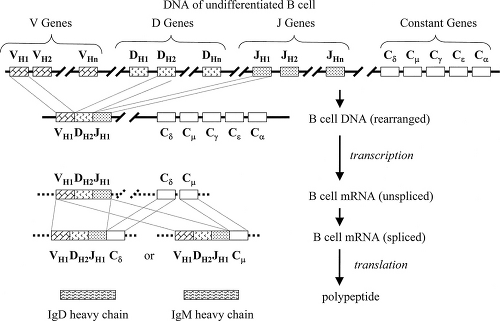
For heavy chains, there are four distinct clusters of genes involved (Fig. 1.4): variable genes (VH), diversity genes (DH), joining genes (JH), and a series of distinct
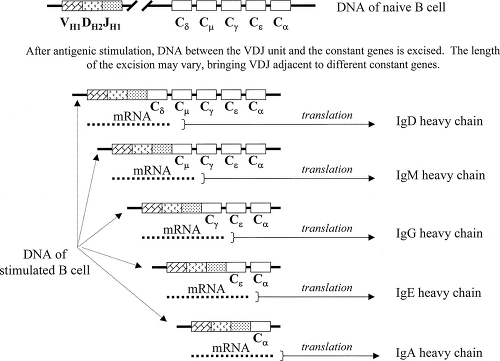
constant genes (Cμ, Cδ, Cγ, Cε, and Cα). As with the light chain genes, each V gene is preceded by a leader sequence (L) that plays a role during synthesis but is subsequently lost. One VH gene, one DH gene, and one JH gene are randomly selected, and the intervening DNA segments are excised and discarded to bring these genes together (VH–DH–JH). Messenger RNA is then transcribed to include both the VH–DH–JH and constant genes, but unlike for the light chains, the processes involving constant genes are distinctly different in stimulated and unstimulated B lymphocytes.
constant genes (Cμ, Cδ, Cγ, Cε, and Cα). As with the light chain genes, each V gene is preceded by a leader sequence (L) that plays a role during synthesis but is subsequently lost. One VH gene, one DH gene, and one JH gene are randomly selected, and the intervening DNA segments are excised and discarded to bring these genes together (VH–DH–JH). Messenger RNA is then transcribed to include both the VH–DH–JH and constant genes, but unlike for the light chains, the processes involving constant genes are distinctly different in stimulated and unstimulated B lymphocytes.
Unstimulated B cells transcribe heavy chain mRNA from VH–DH–JH through the Cμ and Cδ genes. This transcript does not contain the information from the Cγ, Cε, or Cα genes. The mRNA is then spliced to bring VH–DH–JH adjacent to either Cμ or Cδ, which permits the translation of a single continuous polypeptide with a variable domain (from VH–DH–JH) and a constant domain (from either Cμ or Cδ). Thus, the surface immunoglobulin of naïve unstimulated B cells includes only the IgM and IgD isotypes.
After restimulation by antigen (and interaction with certain T lymphocytes, to be described later), previously activated B cells (“memory” B cells) can undergo an isotype switch in which splicing of DNA, rather than RNA, brings the united VH–DH–JH genes adjacent to a constant region gene (3,4,17). This transition is controlled by cytokines secreted by T lymphocytes. Depending on the amount of DNA excised, the VH–DH–JH genes may be joined to any of the different CH genes (Fig. 1.5). As a result of the isotype switch, B-cell “subclones” are generated that produce an array of immunoglobulins that have identical antigen-binding specificity but different isotypes.
Two additional sources of diversity in the variable (antigen-binding) regions of light and heavy immunoglobulin chains occur (3,4,18,19). First, junctional diversity




Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree