Poly (ADP-ribose) Polymerase Inhibitors
Alan Ashworth
INTRODUCTION
Cancer cells may harbor defects in DNA repair pathways leading to genomic instability. This can foster tumorigenesis but also provides a weakness that can be exploited therapeutically. Tumors with compromised ability to repair double-strand DNA breaks by homologous recombination, including those with defects in the BRCA1 and BRCA2 genes, are highly sensitive to blockade of the repair of DNA single-strand breaks, via the inhibition of the enzyme poly(ADP-ribose) (PARP). This provides the basis for a synthetic lethal approach to cancer therapy, which is showing considerable promise in the clinic.
CELLULAR DNA REPAIR PATHWAYS
DNA is continually damaged by environmental exposures and endogenous activities, such as DNA replication and cellular free-radical generation, which cause diverse lesions including base modifications, double-strand breaks (DSB), single-strand breaks (SSB), and intrastrand and interstrand cross-links.1 These aberrations are repaired by distinct repair pathways, which are coordinated to maintain the stability and integrity of the genome. This faithful repair of DNA damage is an essential prerequisite for the maintenance of genomic integrity and cellular and organismal viability. Where one DNA strand is affected and the intact complementary strand is available as a template, the base-excision repair (BER), nucleotide-excision repair, or mismatch repair pathways are used and these pathways are highly efficient at repairing damage. DSBs, more problematic than SSBs because the complementary strand is not available as a template, are repaired by the homologous recombination (HR) or nonhomologous end-joining (NHEJ) pathways.1
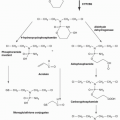
Endogenous base damage, including SSBs, is the most common DNA aberration and it has been estimated that the average cell may repair 10,000 such lesions every day. BER is an important pathway for the repair of SSBs and involves the sensing of the lesion followed by the recruitment of a number of other proteins. PARP-1 (poly[ADP]ribose polymerase) is a critical component of the major “short-patch” BER pathway. PARP is an enzyme, discovered over 40 years ago,2 that produces large branched chains of poly(ADP) ribose (PAR) from NAD+. In humans, there are 17 members of the PARP gene family but most of these are poorly characterized.3,4 The abundant nuclear protein PARP-1 senses and binds to DNA nicks and breaks, resulting in activation of catalytic activity causing poly(ADP)ribosylation of PARP-1 itself as well as other acceptor proteins including histones. This modification may signal the recruitment of other components of DNA repair pathways as well as modify their activity. The highly negatively charged PAR that is produced around the site of damage may also serve as an antirecombinogenic factor. In addition to the BER pathway PARP enzymes have been implicated in numerous cellular pathways.3,4
Two main DSB repair pathways are available within eukaryotic cells: NHEJ and HR.5,6 HR can be further subdivided into the gene conversion (GC) and single-strand annealing (SSA) subpathways.1 Both GC and SSA rely on sequence homology for repair whereas NHEJ uses no, or little, homology.2,3 NHEJ is the most important pathway for the repair of DSBs during G0, G1, and early S phases of the cell cycle, although it is likely active throughout the cell cycle.7,8 This form of DSB repair usually results in changes in DNA sequence at the break site and, occasionally, in the joining of previously unlinked DNA molecules, potentially resulting in gross chromosomal rearrangements such as translocations.9 GC uses a homologous sequence, preferably the sister chromatid, as a template to resynthesize the DNA surrounding the DSB, and therefore generally results in accurate repair of the break. Repair by GC is critically dependent on the recombinase function of RAD51 and is facilitated by a number of other proteins. SSA also involves the use of homologous sequences for the repair of DSBs, but unlike GC, SSA is RAD51-independent and involves the annealing of DNA strands formed after resection at the DSB. The detailed mechanism of SSA is still obscure but it frequently results in the loss of one of the homologous sequences and deletion of the intervening sequence.9 SSA is a potentially important pathway of mutagenesis because a significant fraction of mammalian genomes consist of repetitive elements. GC and SSA are cell-cycle regulated and are most active in S-G2 phases of the cell cycle.10
THE DEVELOPMENT OF PARP INHIBITORS
PARP inhibitors were originally developed as chemopotentiators, which are agents that enhance the effects of DNA damage— a common mechanism of action of drugs used to treat cancer. The rationale was that inhibition of the repair of chemotherapy-induced DNA damage might give greater efficacy. Early studies using relatively nonspecific PARP inhibitors such as 3-aminobenzamide, demonstrated potential synergy with alkylating agents.11 Subsequent studies with more potent PARP inhibitors demonstrated synergy with temozolomide, an observation that was taken into a clinical trial with AG014699,12 a PARP inhibitor developed by Pfizer. This agent is now being developed by Clovis. Although the major focus of this chapter is the use of PARP inhibitors in synthetic lethal therapeutic strategies, their use in chemopotentiation in combination with chemotherapy remains under active investigation, as described later.
BRCA1 AND BRCA2 MUTATIONS AND DNA REPAIR
Heterozygous germline mutations in the BRCA1 and BRCA2 genes confer a high risk of breast (up to 85% lifetime risk) and ovarian (10% to 40%) cancer in addition to a significantly increased risk of pancreatic, prostate, and male breast cancer.13 The genes have been classified as tumor suppressors, because the wild-type BRCA allele is frequently lost in tumors, a phenomenon that occurs by a variety of mechanisms. The BRCA1 and BRCA2 genes encode large proteins that likely function in multiple cellular
pathways, including transcription, cell-cycle regulation, and the maintenance of genome integrity. However, the roles of BRCA1 and BRCA2 in DNA repair have been best documented.14
pathways, including transcription, cell-cycle regulation, and the maintenance of genome integrity. However, the roles of BRCA1 and BRCA2 in DNA repair have been best documented.14
BRCA1- and BRCA2-deficient cells are highly sensitive to ionizing radiation and display chromosomal instability, which is likely to be a direct consequence of unrepaired DNA damage.14 The similar genomic instability in BRCA1- and BRCA2-deficient cells and the interaction of both BRCA1 and BRCA2 with RAD51 suggested a functional link between the three proteins in the RAD51-mediated DNA damage repair process. However, although BRCA2 is directly involved in RAD51-mediated repair, affecting the choice between GC and SSA, BRCA1 acts upstream of these pathways15; both GC and SSA are reduced in BRCA1-deficient cells, placing BRCA1 before the branch point of GC and SSA.15
BRCA1 has a role in signaling DNA damage and cell-cycle checkpoint regulation,14,15 whereas BRCA2 has a more direct role in DNA repair itself. BRCA2 is thought to promote genomic stability through a role in the error-free repair of DSBs by GC via association with RAD51. Aberrations in BRCA2-deficient cells arise at least in part by the use of the SSA pathway. NHEJ, however, is apparently unaffected in BRCA2-deficient cells.14,15 Loss of BRCA2, therefore, results in the repair of DSBs by preferential utilization of an error-prone mechanism, which potentially explains the apparent chromosome instability associated with BRCA2 deficiency.15
The physical interaction between BRCA2 and RAD51 is essential for error-free DSB repair. BRCA2 is required for the localization of RAD51 to sites of DNA damage, where RAD51 forms the nucleoprotein filament required for recombination. The foci of the RAD51 protein are apparent in the nucleus after certain forms of DNA damage and these likely represent sites of repair by HR; BRCA2-deficient cells do not form RAD51 foci in response to DNA damage.15 Two different domains within BRCA2 interact with RAD51, the eight BRC repeats in the central part of the protein and a distinct domain, TR2, at the C-terminus.16
PARP-1 INHIBITION AS A SYNTHETIC LETHAL THERAPEUTIC STRATEGY FOR THE TREATMENT OF BRCA-DEFICIENT CANCERS
Synthetic lethality is defined as the situation when a mutation in either of two genes individually has no effect, but combining the mutations leads to death.17 This effect was first described and studied in genetically tractable organisms such as Drosophila and yeast.17,18 This effect can arise because of a number of different gene-gene interactions. Examples include two genes in separate semiredundant or cooperating pathways, and two genes acting in the same pathway where loss of both critically affects flux through the pathway. The implication is that targeting one of these genes in a cancer where the other is defective should be selectively lethal to the tumor cells but not toxic to the normal cells. In principle, this should lead to a large therapeutic window.19 The original suggestion that the concept of synthetic lethality could be used in the selection or development of cancer therapeutics came from Hartwell et al.,18 and from experiments performed in yeast. Synthetic lethal screens have now been performed in a number of model organisms20 and in human cells,21 and these have revealed multiple potential gene-gene interactions, some of which could be exploited clinically. However, synthetic lethal therapies have not been clinically used until recently, when evidence has been provided for PARP-1 inhibition as a potential synthetic lethal approach for the treatment of BRCA-mutation-associated cancers.
PARP-1 inhibition causes failure of the repair of SSB lesions but does not affect DSB repair.22 However, a persistent DNA SSB encountered by a DNA replication fork will cause stalling of the fork and may result in either fork collapse or the formation of a DSB.23 Therefore, the loss of PARP-1 increases the formation of DNA lesions that might be repaired by GC. As a loss of function of either BRCA1 or BRCA2 impairs GC,14,15 a loss of PARP-1 function in a BRCA1- or BRCA2-defective background could result in the generation of replication-associated DNA lesions normally repaired by sister chromatid exchange. If so, this might lead to cell-cycle arrest and/or cell death. Therefore, PARP inhibitors could be selectively lethal to cells lacking functional BRCA1 or BRCA2 but might be minimally toxic to normal cells. This would indicate a synthetic lethal interaction between PARP and BRCA1 or BRCA2. Exemplifying this principle, potent inhibitors of PARP were applied to cells deficient in either BRCA1 or BRCA2. Cell survival assays showed that cell lines lacking wild-type BRCA1 or BRCA2 were extremely sensitive to these agents compared with heterozygous mutant or wild-type cells.24,25
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree