PARATHYROID HORMONE RECEPTORS
Part of “CHAPTER 51 – PARATHYROID HORMONE“
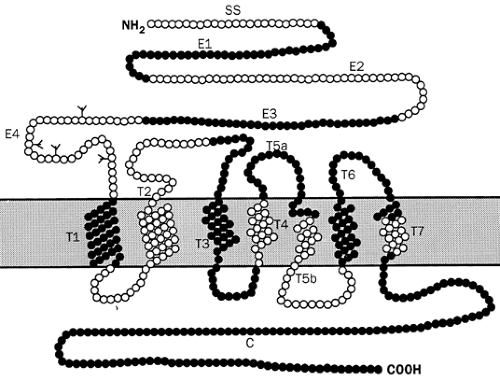
The PTH/PTHrP receptor is a member of the G-protein–linked receptor superfamily. These glycoproteins have hydrophobic sequences that are thought to span the plasma membrane seven times. These receptors couple to intracellular effectors through guanine nucleotide binding regulatory proteins or G proteins, which are heterotrimeric, consisting of alpha, beta, and gamma subunits. In the inactive state, the alpha subunit binds GDP; however, when the receptor is occupied by ligand, the G protein is activated by exchange of GTP for GDP and dissociates into α and β-γ subunits. The alpha subunit, with GTP bound, can then stimulate (Gsα) or inhibit (Giα), the activity of effector molecules such as adenylate cyclase or phospholipase C. The α subunit also has an intrinsic GTPase activity that hydrolyzes GTP to GDP, terminating the effector activation.
The PTH/PTHrP receptor belongs to a subgroup of the G-protein coupled receptor superfamily (group II GPCR) that, by virtue of their structural similarities, also includes the receptors binding secretin, calcitonin, vasoactive intestinal peptide, glucagon, glucagon-like peptide I, and growth hormone–releasing hormone. These receptors show no significant sequence identity with other known G-protein–linked receptors. All of these receptors couple to the adenylate cyclase effector by means of Gsα. The activated PTH/PTHrP receptor couples to at least two effectors: adenylate cyclase and phospholipase C. Other members of this subgroup—including the receptors for calcitonin, glucagon, and glucagon-like peptide I—also couple to multiple signaling pathways.
PTH/PTHrP receptor cDNAs have been cloned and characterized from several species and tissues, including opossum kidney, rat bone, and human kidney and bone.110,111,112 and 113 The amino acid sequences of the receptors are highly homologous, demonstrating a marked conservation across species. The opossum and rat PTH/PTHrP receptors bind NH2-terminal analogs of PTH and PTHrP with similar affinity, while the human receptor appears to bind PTH somewhat preferentially. This is consistent with the study results of relative bioactivities of PTH and PTHrP in vivo in humans, that showed PTHrP 1–34 to be less potent than PTH 1–34.114,114a
The gene for the PTH/PTHrP receptor contains multiple introns (Fig. 51-6). For example, the mouse gene has at least 15 exons—14 of which encode the receptor protein—that span more than 32 kb of genomic DNA.115 There are eight exons containing predicted membrane-spanning domains. These exons are heterogeneous in length, and three of the exon-intron boundaries fall within putative transmembrane sequences, suggesting that these exons did not arise from duplication events. The exon-intron organization of the PTH/PTHrP gene is similar to that of the growth hormone–releasing hormone gene, especially in the transmembrane regions, suggesting that the two genes evolved from a common precursor. PTH/PTHrP receptor gene transcription in mice is controlled by two promoters; P1, which is selectively active in kidney, and P2, which functions in a variety of tissues. Although P1 and P2 are conserved in the human PTH/PTHrP receptor gene, P1 activity in the kidney is weak, and a third promoter, P3, accounts for the majority of renal PTH/PTHrP receptor transcripts in humans.116 During development, only P2 is active at midgestation in many human tissues, including calvaria and long bone. Thus, factors regulating the well-conserved P2 promoter control PTH/PTHrP receptor gene expression during skeletal development. Later in development, receptor gene expression is up-regulated with the induction of both P1 and P3 promoter activities.116a
The PTH/PTHrP receptor gene promoters lack consensus TATA elements, and the downstream promoter P2 and P3 promoters are GC-rich, which would be consistent with the widespread expression of the receptor mRNA. Although the receptor mRNA is highly expressed in kidney and bone, which are the primary target tissues of PTH, it is also expressed in nonclassic target tissues such as liver, brain, smooth muscle, spleen, testis, and skin.112,117 In most of these tissues, the receptor probably mediates the local action of PTHrP. Although the predominant transcript is 2.5 kb, some tissues also express smaller or larger transcripts that probably are the result of alternative splicing of the primary transcript. The functional significance of these different forms of the receptor is unknown.
A second related receptor, PTHR2, which is the product of a distinct gene and binds PTH but not PTHrP, has been identified.118 Its expression is limited to brain, pancreas, testis, and placenta; its function is unknown. Ligand-binding specificity of the PTH/PTHrP and the PTH2 receptors resides predominantly, but not exclusively, in its NH2-terminal extracellular domain, whereas activation and generation of the cAMP signal is generated in the membrane-associated domain and involves specific amino acids in transmembrane domain (TM) 3 and extracellular loop 2. This suggests a two-step interaction of PTH and receptor, whereby the ligand first complexes with the NH2-terminal domain of the receptor, after which the complex interacts with the membrane-associated part of the receptor to generate the signal.
Extensive analysis of the PTHR has also been performed in an attempt to delineate the structural basis of its function. Such studies using mutant and chimeric PTHR have indicated that the NH2-terminal extracellular domain of the PTHR in particular is important for ligand binding and appears to interact with the COOH region of PTH or PTHrP.119 Nevertheless, amino acid residues elsewhere in the PTHR, such as in the third extracellular loop, may also contribute to this binding.120 Residues in the membrane-spanning helices of the PTHR may be required for binding residues in the NH2-terminal domain of the ligand. Amino acids in the extracellular loops and the transmembrane regions are involved in signaling by PTH analogs, and residues in several intracellular loops and the COOH terminal intracellular tail are implicated in linkage to both Gsα and Gqα. Residues in the second intracellular loop appear to be especially critical for interaction with Gqα.121 Other studies have identified sites in the COOH tail of the PTHR that may be important for receptor internalization and phosphorylation.122 Consequently, a great deal of important information has been gleaned and continues to be generated on specific sites in the PTHR that may be necessary for hormone binding, for hormone signaling,
and for PTHR regulation. Nevertheless, a comprehensive understanding of the detailed interaction of PTH and of PTHrP with the PTHR will have to await the determination of the three-dimensional structure of the bimolecular complex.
and for PTHR regulation. Nevertheless, a comprehensive understanding of the detailed interaction of PTH and of PTHrP with the PTHR will have to await the determination of the three-dimensional structure of the bimolecular complex.
High circulating levels of PTH in hyperparathyroid states have been associated with hormonal desensitization in target tissues, apparently caused by diminished receptor capacity and a postreceptor reduction in functional levels of Gs.123,124 PTH receptors, similar to many peptide hormone receptors, appear to be subject to down-regulation. The renal resistance to PTH often seen in hyperparathyroid states may be the partial result of this kind of regulatory mechanism.125 More widespread reductions in Gs are associated with hormone resistance in the disorder pseudohypoparathyroidism type 1a.
The human PTH/PTHrP receptor gene has been localized to chromosome 3p21.1-22112 (and the PTHR2 gene to chromosome 2q33). A search for PTH/PTHrP receptor defects in patients with apparent resistance to endogenous PTH, such as in patients with pseudohypoparathyroidism type 1b, has produced negative results.126,127 These patients have end-organ resistance to PTH without typical features of Albright hereditary osteodystrophy and therefore are thought likely to manifest a defect in PTH/PTHrP receptor expression or function. Because no mutations were identified in the receptor gene in such patients, it was indicated that mutations in genes for other proteins involved in the PTH/PTHrP signaling pathway are most likely responsible for the defects. Linkage to chromosome 20q13.3, which includes the GNAS1 locus encoding Gsα has been established in kindreds with PHP-1b.128 In addition, the genetic defect is imprinted paternally and is therefore inherited in the same fashion as the PTH resistance in kindreds with PHP-1a and/or pseudopseudo-hypoparathyroidism and in a mouse model heterozygous for ablation of the Gnas gene.129 Although the precise nature of mutations causing PHP-1b remains to be elucidated, an understanding of how mutations at a single chromosomal locus cause overlapping and/or distinct phenotypes is likely to be related to (a) the appreciation of the complex nature of the GNAS1 gene that because of its bidirectional imprinting encodes maternally, paternally, and biallelically derived proteins,130 and (b) the subtle cell-specific imprinting of the Gsα transcript. Newly discovered exons upstream of the Gsα exons encode two different proteins, XLαs and NESP55, which are expressed in neuroendocrine cells and are probably important for secretory vesicle formation and function. It therefore is possible that a structural or regulatory mutation within the complex GNAS1 locus could account for PHP-1b, although this remains to be demonstrated.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree