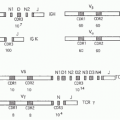
Mouse Ly49
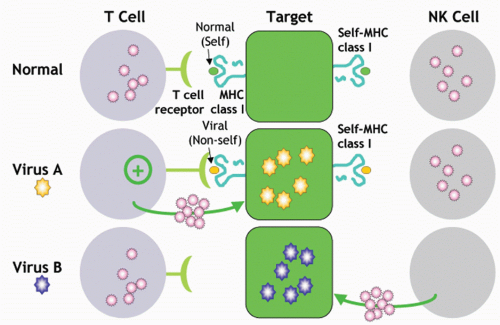
The Ly49A receptor was the first inhibitory MHC-I-specific NK-cell receptor to be described in molecular terms.
191,196 Ly49A was originally identified as a molecule of unknown function on a T-cell tumor.
197,198 It is a disulfide-linked homodimer (44 kDa subunits) with type II membrane orientation, and C-type lectin superfamily homology.
199,200 Previously termed Ly49, it is now appreciated that Ly49A (Klra1) belongs to a family of highly related molecules.
195,201,202,203 Indeed, genetic analysis revealed that the genes for Ly49A and NK1.1 are linked in the NKC
(Fig. 17.3), leading to studies indicating that Ly49A is constitutively expressed on a distinct subpopulation (20%) of NK cells in C57BL/6 mice.
203Multiple lines of evidence indicate that Ly49A is an inhibitory receptor specific for MHC-I, particularly H2D
d: 1) Functional analysis: the Ly49A+ NK-cell subset were equivalent to Ly49A- NK cells in killing several targets, but they could not lyse a large panel of targets that were readily lysed by Ly49A- NK cells. This phenotype was related to MHC-I expression of certain H2 haplotypes on target cells.
191,204,205 Transfected expression of H-2D
d selectively rendered a susceptible target resistant to natural killing by Ly49A+ NK cells. Moreover, killing through disparate stimuli by Ly49A+ NK cells was also inhibited. 2) Cell binding: Ly49A+ tumor cells bound specifically to immobilized MHC-I molecules
206 and to H-2D
d-transfectants.
207 3) Antibody blocking: F(ab′)
2 fragments of mAb directed against either Ly49A or the α1/α2 (but not the α3 domain) of H-2D
d reversed resistance in killing experiments (permitted lysis) and blocked the cell binding assay.
191,204,206,207 4) In vivo expression: the apparent level of Ly49A expressed per NK cell was downregulated in MHC congenic and Tg mice expressing H-2D
d.
208,209,210 This was not due to negative selection because the percentage of Ly49A+ NK cells was unchanged. 5) Gene transfer: primary NK cells and T cells expressing a Ly49A transgene and a Ly49A-transfected NK cell line were specifically inhibited by H-2D
d.
211,212 6) Inhibition by Ly49A is ITIM-dependent, based on gene transfer of mutant Ly49A molecules.
212 7) H2D
d tetramers bind Ly49A transfectants.
213 8) Ly49A tetramers bind H2D
d on transfected cells.
214,215 9) Biophysical studies: recombinant Ly49A binds recombinant H2D
d in surface plasmon resonance (SPR) studies with
KD = ˜2.0 µM.
216 10) Crystallography: the structure of Ly49A complexed with H2D
d was determined.
217 11) Less extensive studies also indicate that Ly49A recognizes H-2D
k, H2D
p, and alleles in H2
r and H2
q.
191,208,213,218,219 Therefore, Ly49A is an MHC-I-specific receptor for H-2D
d, H-2D
k, and H2D
p, and alleles in H2
r and H2
q.
The nature of the Ly49A interaction with MHC-I, however, is fundamentally different from TCR/MHC-I interactions because the former appears to be relatively independent of the specific peptide bound by H2D
d.
220,221 However, bound peptides are required for appropriately folded MHC-I molecules that can be recognized. Despite its structural homology to C-type lectins that are carbohydrate-binding proteins,
222,223 Ly49A does not have the residues for coordinate binding to calcium that is required for lectin binding. Moreover, Ly49A binding to its MHC ligands is not carbohydrate-dependent based on functional, SPR, and crystallographic analyses.
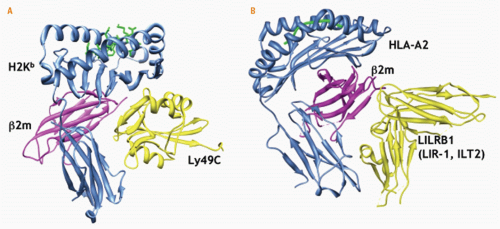
216,224 In the crystallographic structure of Ly49A complexed to H2D
d (2.3Å resolution), the lectin-like structure of Ly49A was confirmed
217 (Fig. 17.4). Two interaction sites were seen between the lectin-like domain of Ly49A and H2D
d: site 1 involved the “left” side of the peptide-binding cleft of H2D
d and a wedge-like site 2 involved the undersurface of the peptide-binding cleft. The residues in Ly49A involved in binding either ligand site are overlapping. Mutational analysis revealed that Ly49A binds site 2 where it contacts α1, α2, and α3 of H2D
d and β2m.
214,216,225 This site is near Asn80, an Asn-linked glycosylation site conserved in all MHC-I molecules, leaving open the issue of whether carbohydrates could affect the interaction, such as affinities or kinetic parameters, but this has not been studied in depth. These studies also provide a structural explanation for species-specific β2m requirements as revealed by functional studies.
226 Thus, Ly49A recognizes site 2 in the MHC molecule in terms of
trans recognition between the NK-cell receptor and target cell MHC-I molecule.
Most but not all other Ly49 receptors are MHCI-specific inhibitory receptors. First noted by Southern blot analysis and cDNA cloning, genome sequence analysis revealed 15 complete Ly49 genes in C57BL/6 mice.
199,200,201,202,227 There is evidence for alternative splicing and alternative transcriptional start sites for the Ly49 genes, though their importance has not been elucidated.
202,228,229 Notably, Ly49C has broad specificity for H2 alleles, as revealed by tetramer staining, and is the only known inhibitory NKcell receptor specific for an H2
b haplotype allele (H2K
b) in C57BL/6 mice, notwithstanding unconfirmed reports that Ly49I also binds H2K
b.
168,213,230 Ly49C is recognized by two mAbs: 5E6, which also binds Ly49I, and 4LO3311, which has exquisite specificity for Ly49C.
231,232,233 X-ray crystallographic studies have indicated that Ly49C binds H2K
b in a manner similar to Ly49A interaction with H2D
d (see
Fig. 17.4).
234,235 Only a site 2 interaction was seen with the contact residues, showing a similar but distinct topology to the Ly49A-H2D
d interaction. Interestingly, however, peptides bound to H2K
b clearly affect functional interactions with Ly49C
236 and affinities as measured by SPR.
234 However, Ly49C does not directly engage the peptide, indicating long-range effects. Finally, both Ly49A and Ly49C
appear to undergo conformational changes upon ligand binding, and a Ly49 dimer can engage two MHC-I molecules.
235,237 Thus, Ly49 receptors bind their MHC ligands in a structurally related manner.
Recent studies suggest that Ly49 molecules also can bind MHC in a
cis interaction between receptor and ligand on the NK cell itself.
168,238 For example, an MHC ligand for Ly49A or Ly49C on the same cell prevents binding of MHC tetramer.
168,238 If the cells are briefly exposed to mild acidic conditions, MHC-I expression is lost (due to disruption of the noncovalently linked MHC-I heterotrimer), and Ly49A binding to cognate MHC-tetramers is restored. This
cis interaction is dependent on site 2 residues in the MHC molecule. These findings may help explain the observation that the presence of self-MHC ligands leads to downregulation of Ly49 expression, as previously noted on primary NK cells in MHC congenic mice.
208,209,210 Cis interactions may also explain functional differences in NK cells in MHC-congenic mice or NK cells that do or do not express MHC ligands in Tg mice that are mosaic for MHC expression.
239,240,241 Finally, there are biophysical data supporting a role for the relatively long, flexible stalk region of Ly49 receptors in allowing either
trans or
cis interactions.
242 At the moment, the physiologic importance of
cis interactions is incompletely understood but may be relevant to NK-cell tolerance and education, as discussed below.
Although they have not been studied as extensively as Ly49A and Ly49C, other inhibitory Ly49 receptors and their ligands have been identified
202,230 and characterized with other MHC allele specificities, as detailed in a recent review.
243 Moreover, they are structurally related.
235,244 Individual NK cells may express multiple Ly49 receptors simultaneously,
210,233,245 often (but not always) two or more, suggesting that individual NK cells may be inhibited by more than one MHC-I molecule.
Ontogenetic studies demonstrate that the total repertoire of Ly49 expression does not reach adult levels until sometime after 3 weeks of age, concomitant with attainment of full NK-cell cytolytic activity.
246 Thereafter, the expression of Ly49 receptors is generally thought to be fixed and stable on an individual NK cell. Ly49E is expressed only on fetal NK cells, but NK cells in mice deficient in Ly49E are otherwise normal.
247 In adult mice, developmental studies indicate that Ly49 receptors are first expressed on immature NK cells in the BM, before a phase of constitutive proliferation
98 that appears to be modestly affected by MHC haplotype.
168 There are only modest effects on the final “repertoire” of MHCspecific receptors expressed by splenic NK cells in different MHC environments.
210,248The expression of inhibitory Ly49 receptors appears to occur in a stochastic manner. There is evidence for monoallelic expression of Ly49 receptors (expression from one chromosome), initially described as “allelic exclusion,” a term that has fallen out of favor because it has a specific meaning and mechanism for TCRs and BCRs.
249 At least some Ly49 genes possess bidirectional, overlapping promoters directed in opposite orientations.
250 Transcription factors driving transcription in one direction prevent binding of other factors driving transcription in the opposite direction. Directionality and monoallelic expression may also be controlled by DNA methylation.
251 A “probabilistic” model has been proposed to explain these findings that may also explain the stochastic expression of Ly49 genes and their stable expression. However, recent studies of
Ly49 indicate highly variable transcriptional start sites, suggesting that the probabilistic model may not be correct.
229 Other data indicate that TCF-1 but not LEF-1 in the T-cell factor/lymphoid enhancer family of DNA-binding proteins affects some but not all Ly49 receptor expression.
252,253,254 Thus, the elements controlling Ly49 gene expression are incompletely understood.
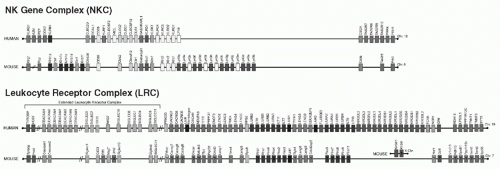
Analysis of the Ly49 receptors thus far is largely based on examination of the C57BL/6 alleles, but the Ly49 receptors display extensive polymorphism. The Ly49 family is encoded in the NKC located on mouse chromosome 6 with the syntenic human region being chromosome 12p13.2
195,203,255,256 (see
Fig. 17.3). While the NKC also contains genes for other lectin-like receptors, the Ly49 genes are clustered with the exception of
Ly49b. Corresponding to restriction fragment length polymorphic (RFLP) variants originally detected with the Ly49A cDNA,
203 there is significant allelic polymorphism of the Ly49 cluster between inbred mouse strains with differences in gene number as well as alleles for the
Ly49 genes.
227,257,258,259 In contrast to C57BL/6J mice, genomic sequence analysis shows 8 putative
Ly49 genes in BALB/c mice, 19 in 129 mice (of which at least 9 appear to be pseudogenes) and 22 in NOD mice. Array-based comparative genomic hybridization analysis of 21 mouse strains compared to the reference C57BL/6J strain indicated that these mice could be grouped into five clusters that correspond to or are predictive of restriction fragment length polymorphic patterns on Southern blot analysis.
203,260 There are also multiple alleles for individual
Ly49 family members.
227,249,257,258,261 Thus, there is significant polymorphism of the
Ly49 molecules at both the haplotype (gene numbers) and individual gene (alleles) levels, not unexpected because the Ly49 molecules bind highly polymorphic MHC-I molecules.
The MHC-I specificities have generally been well characterized for only a few Ly49 alleles. Interestingly, mAbs specific for one Ly49 allele may bind another molecule with a different function or specificity in another mouse strain,
262,263,264 similar to what was recognized for mAb reactivity with different MHC alleles.
265 Thus, the polymorphisms also raise practical issues when studying Ly49 molecules in different mouse strains.
Finally, it should be noted that Ly49 receptors may be expressed by other cells; some are selectively expressed on non-NK cells and some may have specificities for non-MHC ligands. NKT and other T-cell subsets may express Ly49 receptors but they have not been thoroughly studied.
266,267 Ly49B and Ly49Q are not expressed on NK cells; rather, they are expressed on myeloid cells.
268,269 Interestingly, Ly49Q recognizes H2K
b and positively regulates TLR signaling.
270,271 On the other hand, Ly49E appears to recognize urokinase plasminogen activator, though physical binding has not been established.
272 Interestingly, Ly49B, Ly49E, and Ly49Q are predicted to be distinct in fine structure from the
known MHC-I-specific Ly49 receptors on NK cells.
235 Thus, while Ly49 receptors are predominantly NK-cell inhibitory receptors for MHC class I, they may have other roles that are less well understood; still other Ly49 receptors have activation function, as discussed subsequently.
Human Killer Immunoglobulin-like Receptors
In contrast to mouse NK cells, human NK cells can be cloned by limiting dilution in the presence of irradiated feeder cells, phytohemagglutinin, and IL-2, leading to establishment of short-term NK-cell clones that have differences in target killing and surface molecules. mAbs were isolated that reacted specifically with these clones; reactivity correlated with the capacity of the clones to kill certain tumors, and the mAbs affected cytotoxicity. This general approach led to the identification of the human NK-cell receptors.
A series of studies
273,274,275 showed that the mAbs GL183 and EB6 identify serologically distinct 55 kDa or 58 kDa molecules, initially termed p58. These molecules had several features: 1) selective expression on overlapping NK cell subsets; 2) expression on NK cell clones correlated with expression of certain human leukocyte antigen (HLA) class I alleles on resistant targets; 3) a target susceptible to a given NK-cell clone bearing p58 molecules reactive with either mAb was made resistant by transfection of cDNAs encoding certain HLA-C molecules; 4) the otherwise resistant, HLA-C-transfected targets could be lysed in the presence of the appropriate anti-p58 mAbs. The mAb effect occurred with F(ab′)
2 fragments, suggesting that the interaction between p58 and an HLA class I molecule on the target cell inhibits the NK cell. Thus, the p58 molecules displayed features consistent with a role as inhibitory human NK-cell receptors specific for MHC-I, analogous to the mouse Ly49A receptor that was being studied in parallel, as described previously.
Other studies noted that NK-cell specificity was skewed when the NK cells were grown in the presence of cells bearing allo-MHC determinants.
276,277 This specificity correlated with reactivities that mapped to paired residues at position 77 and 80 in the .α1 domain of HLA-C. All known HLA-C molecules could be divided into two groups, one with Asn77-Lys80 (HLA-Cw2, -Cw4, -Cw5, -Cw6) and the other with Ser77-Asn80 (HLA-Cw1, -Cw3, -Cw7, -Cw8). Indeed, transfection analysis showed that p58 specificity for HLA-C molecules was related to expression of the EB6 epitope for the former (specificity 1, now termed HLA-C1), whereas the latter was related to the GL183 epitope (specificity 2, HLAC2) on the NK-cell clones.
276,278,279 Thus, human NK-cell receptors showed promiscuous specificity that was dependent on residues 77 and 80 in HLA-C.
The NKB1 (p70) molecule was serologically similar to p58 molecules with regard to subset expression, and correlation of expression on NK-cell clones to specificity for HLA class I.
280,281 In contrast to p58 molecules, however, NKB1 had a distinct M
r (70 kDa) and specificity for HLA-B. The NKB1+ clones were specifically inhibited by targets expressing transfected HLA-Bw4 molecules, and the anti-NKB1 mAb reversed the inhibition. Analysis of informative HLA-B alleles showed that this specificity was conferred by a region in the α1 domain overlapping the area on HLA-C recognized by p58 molecules.
282 Finally, HLA-A3, -A11-specific receptors have similar properties to p58 and NKB1 except that they appear to be disulfide-linked dimers termed p140,
283 whereas others have found that a monomeric HLA-A3-specific receptor resembles NKB1.
284 Thus, representative alleles of all classical HLA class I loci are capable of inhibiting NK cells through p58/NKB1/p140 receptors although HLA-B and -C alleles dominate human NK-cell specificities, and it is not yet known if there are receptors reactive with each HLA allele.
When the cDNAs for the p58 and NKB1 molecules were cloned, they were surprisingly found to encode type I integral membrane proteins with Ig-like domains
285,286,287 unlike the lectin-like Ly49 family of type II receptors. The Ig-like receptors are now collectively known as killer Iq-like receptors (KIRs) or CD158.
288,289 The KIR nomenclature is based on whether the receptor has two or three Ig-like external domains (KIR2D or KIR3D, respectively), and possession of a long (L) or short (S) cytoplasmic domain. In general, the L forms are inhibitory because they contain ITIMs, whereas the S forms appear to be activation receptors (see following discussion). Each distinct receptor is also designated by a number. The KIR2DL1 (CD158a, p58.1) molecule bears the original EB6 epitope and is specific for HLA-C (Lys80, specificity 2), whereas KIR2DL2 (CD158b1, p58.2) and KIR2DL3 (CD158b2, p58) have the GL183 epitope and are specific for HLA-C (Asn80, specificity 1). (As detailed in the following, structural analysis supports grouping of HLA-C alleles into two mutually exclusive groups, HLA-C1 and HLA-C2, based on direct interaction of KIR2DL2/3 and KIR2DL1, respectively, with residue 80 of HLA-C, validating original functional groupings but simplifying HLA-C groupings to just residue 80.
276,277) KIR2DL4 (CD158d, p49) reportedly binds HLA-G
290 but displays both inhibitory and activation functions.
291,292,293 KIR3DL1 (CD158e1, NKB1, NKAT3) is specific for HLA-A and HLA-B molecules with the Bw4 epitope.
282 KIR3DL2 (CD158k, p140, NKAT4) has HLA-A3 and HLA-A11 specificity.
283,284There is unequivocal evidence that the KIR2DL and KIR3DL molecules are inhibitory HLA class I-specific receptors. In addition to the data with NK cell clones and mAbs mentioned previously, the following have been described: 1) KIR bind directly to HLA class I: soluble KIR2DL-Fc fusion proteins bind cells expressing the appropriate transfected HLA class I alleles.
294,295 In addition, a soluble KIR2DL molecule containing only the extracellular domain binds specifically to its HLA-C ligand in solution.
296 2) Gene transfer of KIR: KIR2DL specificity and inhibitory function were transferred when KIR2DL cDNAs were transiently expressed with vaccinia constructs in human NK-cell clones.
294 Similarly, Tg expression of KIR2DL2 in mice conferred inhibition of rejection of BM expressing Tg HLA-Cw3.
297 3) SPR measurements indicate that the KIRs bind their HLA ligands with
Kd = ˜10 µM.
298,299,300,301 Binding is affected by peptide bound by HLA molecule.
300,301 Through histidine-rich domains, the KIRs bind Zn++, which affects KIR multimerization and binding kinetics to HLA ligands.
302,303 4) Crystallographic studies demonstrate
KIR2DL1 (2.8 Å resolution) and KIR2DL2 (3.0 Å resolution) interactions with their cognate HLA ligands.
299,304 Thus, KIR molecules are clearly MHC-I-specific inhibitory receptors on NK cells.
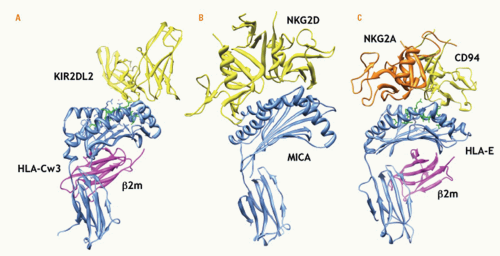
Interestingly, structural studies indicate that KIR molecules bind HLA class I molecules in a manner analogous to recognition of MHC by TCRs
(Fig. 17.5). In particular, both KIR2DL1 and KIR2DL2 use surface loops near their interdomain hinge regions to bind their cognate HLA-C ligands (Cw4 and Cw3, respectively) with a footprint overlying the “right” side of the peptide-binding cleft (when viewed from the “top” in standard depictions of MHC-I molecules).
299,304 The receptors bind both α1 and α2 helices with interactions between KIR2DL1 and Lys80 of HLA-Cw4 and between KIR2DL2 and Asn80 of HLA-Cw3. These interactions with residue 80 of the HLA-C molecules were lost when the reciprocal residues were swapped, accounting for the previously described HLA-C groupings and KIR specificities in functional studies
276,277 and mutational analysis indicating that residue 80 is more significant for KIR interaction than residue 77.
305,306,307,308 Although neither KIR2DL molecule has extensive contacts with peptides bound to HLA-C, KIR2DL interactions with HLA-C imposes physical constraints on the p8 position of the peptide. This may account for observed peptide preferences in functional studies and antagonism of certain peptides on KIR inhibitory function.
309,310 A recent structure (1.8 Å resolution) of KIR3DL1 complexed to HLA-B*5701 revealed a similar recognition strategy.
301 Thus, KIRs and Ly49s bind their MHC ligands in markedly different ways, despite their analogous functions as MHC-specific inhibitory receptors.
Also unlike the Ly49s, the KIRs are encoded in the LRC on human chromosome 19q13.4 that encodes many other Ig-like receptors (see
Fig. 17.3)
311; the KIR genes are clustered toward the telomeric end of the LRC.
312 Interestingly, the mouse LRC on chromosome 7qA1 does not include genes for KIR-like molecules that instead are encoded on the X chromosome.
313,314 On the other hand, like the Ly49s, the KIRs display remarkable polymorphism with at least 11 genes
315,316,317 (see
www.ebi.ac.uk/ipd/kir/index.html for updated database).
The human
KIR locus also demonstrates considerable haplotype diversity with at least 27 different haplotypes,
315,316,318 recently defined at the sequence level.
317 While there has not been a consensus definition, two major types of haplotypes have been described.
289 Group B contains one or more of the following:
KIR2DL5,
KIR2DS1,
KIR2DS2,
KIR2DS3,
KIR2DS5, and
KIR3DS1, whereas group A haplotypes have none of these genes. Reflecting extensive allelic polymorphism of individual genes, a large number of different KIR genotypes have been described, and they are distributed differently in the various ethnic populations.
As with the Ly49s, only a few KIR alleles have been well characterized with respect to HLA class I specificities. Nonetheless, these genetic variants have not only provided clues to new receptors and ligand specificities but also valuable links to the role of NK cells and their receptors in
disease pathogenesis (discussed in the subsequent clinical section), and more broadly, human evolution.
Convergent Evolution of Major Histocompatibility Complex-Specific Natural Killer-Cell Receptors
Despite the controversy surrounding the initial cloning of mouse Ly49s and human KIRs and leukocyte Ig-like receptors (LILRs), additional data have provided new interpretations of the distinctly different receptors used by mouse and human NK cells, respectively, to recognize MHC-I. Detailed genome sequence information is available on the NKC and LRC in mice, humans, and other species. In the mouse, MHC-specific NK-cell receptors with Ig-like domains have not yet been described, although there is conservation of several genes in the broader LRC on mouse chromosome 7
312 (see
Fig. 17.3). Activated mouse NK cells do express gp49b (Lilrb4), an Ig-like inhibitory receptor also expressed on other leukocytes, including mast cells.
319,320,321,322 However, it is not expressed by resting NK cells and is not specific for MHC-I. Instead, it binds the integrin αvβ3 and appears more important for responses of other cells, such as neutrophil, eosinophil, and DCs.
323,324,325,326,327,328 Mouse
Kir3dl1 is on the X chromosome (see
Fig. 17.3) and expressed in NK and T cells, but its function and ligand remain unknown.
313,314 The Ly49 locus in humans consists only of
LY49L (
KLRA1) that is a pseudogene because of a point mutation that gives rise to a splicing abnormality.
329 Thus, current data indicate that mouse NK cells do not express functional KIR orthologues while human NK cells do not express functional Ly49 orthologues.
One reason for this discrepancy may be that the corresponding orthologue is present in the genome but has not been identified. Indeed, genomic sequencing has revealed a multitude of candidate orphan receptors in the genome and specifically the LRC and NKC that have yet to be studied carefully.
311,330 In that regard, identification and functional analyses of NK cell receptors led to the recognition that they are encoded in genomic regions containing gene clusters for related receptors that are expressed on other leukocytes, not just NK cells. Dissection of the expression and function of these receptors is a rich area of research that is beyond the scope of this chapter.
The alternative and currently favored view for the discrepancy is that mice and humans independently evolved analogous receptors to serve the same function. While both human and mouse NK cells express a conserved relatively nonpolymorphic lectin-like receptor, CD94/NKG2, it does not possess many of the features that are shared by mouse Ly49 and human KIRs:
both Ly49s and KIRs are constitutively and selectively expressed on naïve, unstimulated NK cells (with exceptions for rare populations of T cells);
both bind MHC-I molecules with intermediate affinity (KD = 2-10 µM);
binding to MHC is promiscuous;
MHC-bound peptides have only a modest effect, if at all, on recognition;
both use ITIMs to inhibit NK-cell activation;
they are expressed in a stochastic fashion on overlapping subsets of NK cells;
a single NK cell simultaneously expresses one or more of either type of inhibitory receptor (each may be functional);
once they are expressed, their expression appears to be stable;
both are germ-line encoded by small families of genes that are clustered in the genome;
both display impressive polymorphism, in terms of gene number and alleles for each gene;
both are related to molecules that lack ITIMs and instead are activation receptors; and
both are involved in NK-cell education by self-MHCI (see following discussion).
Thus, the mouse Ly49 receptors and human KIRs are analogous receptors in an apparently striking example of convergent evolution,
331 whereby each species came up with a different genetic solution to provide extremely important functions for species reproduction and survival.
In other species,
Ly49 and
KIR genes have been analyzed primarily with respect to sequence and gene number.
332 For example, the
LY49L gene in baboons appears to be functional but the putative polypeptide lacks an ITIM.
333 In rats, the
Ly49 cluster appears to have markedly expanded with at least 25 genes,
334,335 demonstrating one of the most rapid rates of gene expansion.
336 Dog, cat, and pig appear to have only one
Ly49, whereas horse represents the only known nonrodent mammal with several
Ly49 genes.
337 The chicken genome has several lectin-like receptor genes that are genetically linked to the MHC.
338,339,340 On the other hand, multiple KIR genes have been described in primates and cattle.
341,342,343,344 Rhesus macaques have a profound plasticity of KIRs with multiple genotypes and haplotypes
345 encoding receptors that bind MHC-I.
346,347 In rat, a KIRlike sequence has been reported,
314 and dogs and cats lack functional KIRs.
348 Interestingly, pigs and marine carnivores each possess a single Ly49 and KIR gene, but it is not clear if these are functional.
348 Finally, several species have no readily identifiable Ly49 or KIR genes
332 (eg, teleost fish instead possess a large number of novel immunetype receptors with sequence homology to mammalian LRC-encoded receptors
349). Perhaps these species have had alternative convergent evolutionary stratetgies to preserve inhibitory MHC-I-specific receptors.
Additional studies of the Ly49-like and KIR-like molecules as well as potential new orthologues in other species will be of interest to evolutionary biologists for several reasons including prior description of NK-like cells in lower vertebrates and MHC genes that are coevolving.
332,350 Moreover, in primitive chordates, NK-like, missing-self-like recognition affects histocompatiblity reactions
351 but the molecular determinants of histocompatibility involves molecules unrelated to mammalian Ly49, KIR, or even MHC itself.
352,353,354 Thus, evolutionary studies of NK-cell receptors and their ligands may provide unique insight into missingself and other histocompatiblity reactions.
While we have focused thus far on mouse Ly49 and human KIR as the major MHC-I-specific receptors, there are other well-described NK-cell receptors belonging to
either the lectin-like receptor or Ig-like receptor superfamilies. These receptors also have specificities for MHC-I molecules.
Human and Mouse CD94/NKG2
The analysis of CD94 (Klrd1) and NKG2 (Klrc, excluding NKG2D [Klrk1]) family of molecules was especially challenging and required insightful investigations. Identified by subtractive hybridization, the human NKG2 molecules are type II integral membrane proteins with external C-type lectin domains
355 encoded in the NKC
356 (see
Fig. 17.3). Initial attempts to express NKG2 molecules on the cell surface were thwarted. Meanwhile, mAb reactivity suggested that CD94 was variably expressed on human NK cells as a disulfidelinked dimer (70 kDa NR, 43 kDa R),
357 and both activation and inhibition functions for CD94 were described.
358,359,360,361 Surprisingly, cDNA cloning revealed that CD94 has a short seven amino acid cytoplasmic domain, suggesting that it cannot signal on its own.
360 Furthermore, anti-CD94 immunoprecipitates were not detectable from radiolabeled CD94 transfectants, despite easily detectable expression on FACS analysis with the same mAbs.
362,363 These apparent discrepancies were resolved when it became clear that CD94 heterodimerizes with NKG2 molecules
362; NKG2A is the 43 kDa molecule previously identified as Kp43 with anti-CD94 mAbs.
364,365 While CD94 may be expressed as a homodimer, the NKG2 partner provides the signaling motif, whether activation or inhibition.
364,366 (NKG2B is an alternatively spliced form of NKG2A. The rest of the NKG2 family is discussed in the following.)
The ligand specificity for CD94/NKG2 receptors was also initially thought to be promiscuous as interactions with many classical (class Ia) and nonclassical (class Ib) HLA molecules had been described.
359,362,364,367,368,369,370 However, human CD94/NKG2 receptors directly recognize HLA-E, a MHC-Ib molecule homologous to mouse Qa-1.
371,372,373 HLA-E (and Qa-1) is widely expressed with limited polymorphism.
374,375,376 While HLA-E heavy chain is expressed with β2m and a peptide occupying its peptide-binding cleft, its peptide repertoire is largely derived from the leader sequences of MHC-Ia molecules, as previously noted for mouse Qa-1.
377,378 HLA-E (or Qa-1) expression thus requires normal production of HLA-E (or Qa-1) and synthesis of certain MHC-Ia molecules. Mouse CD94/NKG2 recognizes Qa-1 that shares many features with HLA-E.
379,380 These findings need to be considered in the context of the prevailing view at the time that mouse and human NK cells use structurally different receptors to recognize MHC-I molecules.
381 Clearly, the CD94/NKG2 receptors and their ligands are conserved in humans and mice.
The crystal structure of human CD94/NKG2A bound to HLA-E was resolved to 2.5Å and 4.4Å resolution.
382,383 Remarkably, CD94/NKG2A interfaces with HLA-E in a manner analogous to TCR recognition of peptide-loaded MHC-I,
384 including TCR binding to HLA-E itself.
385 Both innate and adaptive receptors for HLA-E lay across the peptide-binding cleft, though peptide itself plays a relatively minor role in binding CD94/NKG2A.
382,383 Strikingly, this binding is distinct from that of Ly49 receptors to their classical MHC-I ligands (see previous discussion). On the other hand, this binding interface is similar to binding of another NKC-encoded, lectin-like receptor, the NKG2D activation receptor, to its MHC-like ligands (see following discussion). Thus, NKC-encoded, lectin-like receptors surprisingly use different strategies to contact their ligands, even though these ligands have structurally related MHC-I folds.
Despite its conservation between mice and humans, the role of CD94/NKG2 receptors in NK-cell function is still incompletely understood. For example, viruses encode peptides that bind and enhance expression of HLA-E, providing a CD94/NKG2A-dependent mechanism to avoid NK-cell attack.
386,387 By contrast, human NK cells expressing CD94/NKG2C (an activation receptor) expand in response to cytomegalovirus (CMV)-infected targets.
388 When a large number of human NK-cell clones were obtained from two normal individuals, CD94/NKG2 seemed to account for the majority of self-MHC-specific receptors on clones from one individual whereas KIRs dominated the self-specific receptors on clones from the other individual, suggesting that some individuals may depend on CD94/NKG2 for self-tolerance.
389 Qa-1 and HLA-E can present peptides derived from other molecules, including the signal sequence of heat shock protein 60 (Hsp60) that is induced by a number of stimuli,
390,391 a multidrug resistance transporter,
392 or blastocyst MHC expressed in embryonic tissues.
393 This may result in loss or gain of recognition by the inhibitory CD94/NKG2A receptor, suggesting intrinsic mechanisms to perturb inhibition by CD94/NKG2A in certain circumstances.
Yet, CD94 appears to be dispensable in certain strains of mice, such as DBA/2J, that do not appear to have any untoward NK-cell phenotype.
394 This finding has been recapitulated in studies of a CD94 knockout mouse on the 129 strain background.
395 Interestingly, CD94 knockout mice are more susceptible to ectromelia virus,
396 as detailed below.
CD94/NKG2 molecules may be important in T-cell function. CD94/NKG2A is rapidly induced on antigenspecific CD8+ T cells during polyoma virus and other infections,
397,398,399 and CD8+ T cells expressing CD94/NKG2A preferentially proliferate during persistent infection, suggesting that CD94/NKG2 receptors may play a role in memory T-cell responses,
400 and that TCR specificity is correlated with CD94/NKG2A expression by human CTL.
401 Indeed, CD94/NKG2A inhibits antigen-specific cytotoxicity in polyoma virus responses, although this effect is pathogen-dependent. CD94/NKG2A has been studied with other viral infections, including herpes simplex virus,
402 murine CMV,
403 gHV68, and influenza,
404 for example. Although not all studies revealed functional consequences, the recurring theme is the appearance of CD94/NKG2A on previously activated CD8+ T cells. Thus, CD94/NKG2 receptors may regulate T-cell responses.
Other Human Immunoglobulin-like Receptors Specific for Major Histocompatibility Complex-I Molecules
The human LILR family is encoded in the LRC (see
Fig. 17.3), just centromeric to the
KIR genes. There are two general forms of these receptors, subfamily A that appears to be activation receptors, and subfamily B that has the ITIMs
characteristic of inhibitory receptors. The best characterized members, LILRB1 (also known as CD85j, Ig-like transcript 2 [ILT2], or leukocyte IG-like receptor 1 [LILR1]) is broadly expressed whereas LILRB2 (CD85d, ILT4, or LIR2) is not expressed on NK cells but is expressed by myelomonocytic cells, including DCs and monocytes. Both recognize HLA class I molecules
405,406 with LILRB1 exclusively binding folded HLA class I molecules with β2m, whereas LILRB2 can bind both folded and free HLA class I heavy chains.
407 Interestingly, a human CMV protein, UL18, binds LILRB1 with 1000-fold higher affinity than HLA molecules, implicating a role for LILRB1 in host defense.
408 The ligands and functions of other LILRBs (LILRB3 [CD85a, ILT5, LIR3], LILRB4 [CD85k, ILT3, LIR5], LILRB5 [CD85c, LIR8], and LILRB6 [CD85b]) are as yet unknown but they may not be able to bind HLA due to structural constraints.
409LILRB1 has four Ig-like domains and binds a conserved region in the α3 domain of most, if not all, classical and nonclassical HLA class I molecules (HLA-A, -B, -C, -E, -F, and -G).
408 Interestingly, the crystal structure of LILRB1 bound to HLA-A2 (3.4Å resolution) reveals that it binds MHC molecules under the peptide-binding domain where it contacts α3 and β2m, more akin to Ly49 engagement of MHC than KIR
409 (see
Figs. 17.4 and
17.5). Even though LILRB1 and LILRB2 have differing capacities to bind free HLA molecules, LILRB2 has an HLA class I binding site that overlaps with but is distinct from that for LILRB1.
410 LILRB1 binds UL18 in a manner structurally similar to HLA class I binding.
411Interestingly, LILR genes demonstrate allelic polymorphisms,
412 though less so than the adjacent KIR cluster.
413 Nonetheless, polymorphisms in LILRB1 may affect receptor expression.
414 Moreover, polymorphisms in LILR genes are associated with certain autoimmune diseases, such as rheumatoid arthritis.
415
Major Histocompatibility Complex-Independent Natural Killer-Cell Inhibitory Receptors
As already mentioned, NK cells also express inhibitory receptors for non-MHC ligands, such as mouse gp49b, which binds the αvβ3 integrin,
323 and there is a growing list of other such molecules.
416 Some of these receptors will be discussed in a more appropriate context in the following sections. Most of these receptors contain cytoplasmic ITIMs so their inhibitory function can be predicted even if not directly tested, though some caution is required because the motifs may be involved in other signaling processes.
Human and mouse NK cells express the ITIM-bearing leukocyte-associated Ig-like receptor 1 (LAIR-1, CD305), which is an Ig-like molecule broadly expressed by most leukocytes and encoded in the LRC.
417,418 Initial reports indicating that LAIR-1 binds epithelial cellular adhesion molecule were irreproducible.
419,420 Instead, LAIR-1 binds multiple forms of collagen,
421 which has been validated in crystallographic and biochemical studies.
422 Interestingly, LAIR-1 mediates inhibition that is independent of Src homology 2 (SH2)-domain-containing phosphatases, and instead recruits C-terminal Src kinase (Csk),
423 suggesting that Csk may be involved in inhibitory signaling. Although the in vivo context for functional interaction awaits further characterization, it is reminiscent of the broader reactivity of the Siglecs.
The CD33-related sialic acid binding Ig-like lectins (CD33rSiglecs) are type I receptors with varying numbers of Ig-like domains expressed on a broad array of cells and encoded in the “extended” LRC
312,424,425 (see
Fig. 17.3). Despite having sialic acid recognition in common, the Siglecs appear to show differences in carbohydrate recognition, depending on the specific glycan context.
424 Human NK cells express Siglec-7 (p75, adhesion inhibitory receptor 1 [AIRM1]) and Siglec-10, whereas some mouse NK cells express a related Siglec-E.
425 Siglec-7 has been most extensively studied. As expected, its cytoplasmic ITIM can recruit SHP-1 and inhibit NK-cell functions.
426,427 Moreover, expression of its ligand on targets inhibits NK cells in a Siglec-dependent manner.
428 However, the effects appear to be modulated by
cis interactions between the Siglec receptor and its carbohydrate ligands on the NK cell itself,
424,428,429 reminiscent of
cis interactions between Ly49 and MHC ligands, as previously discussed.
The Ig-like receptors, termed paired Ig-like receptor (PILR)α and β, are not encoded in the LRC, rather on human chromosome 7 (mouse chromosome 5).
430,431 Mouse PILRα is an ITIM-containing inhibitory receptor, whereas PILRβ is an activation receptor that couples to DAP12.
430,432 Both receptors are expressed on NK (and other immune) cells and recognize CD99. Interestingly, sialylated O-linked glycans on CD99 are involved in recognition by PILRs.
433Thus, NK cells (and other leukocytes) express multiple inhibitory receptors that are capable of MHC-independent recognition. How they participate in NK-cell responses and contribute to MHC-dependent effects are beginning to be elucidated, and some appear to play a specific role in the context of activation receptors, as discussed in the following.