Molecular Diagnostics of Lung Cancer
Sanja Dacic
The diagnosis of lung carcinoma is largely based on morphology. Ancillary tools such as histochemical stains and immunohistochemistry are used in occasional cases. Our understanding of lung carcinoma carcinogenesis at the genetic level has significantly expanded over the last few decades, but only several observations have been implemented into molecular diagnostics of lung cancer. Development of invasive lung carcinoma is a result of sequential accumulation of genetic and epigenetic changes which are also reflected at the morphological level. However, every tumor does not harbor the same mutations in the same genes, and the different tumors acquire mutations in a different sequence. This represents a fundamental limitation in diagnostic sensitivity and specificity of genetic analysis regardless of the testing approach. This chapter will focus on applications of molecular assays that are currently used to address some diagnostically problematic areas including differentiation between primary and metastatic tumors, origin of tumors occurring in lung allografts, diagnosis of salivary gland tumors, and potential classification of poorly differentiated carcinomas.
PRIMARY VERSUS METASTATIC TUMORS
The distinction between a primary lung carcinoma and lung metastasis in patients with prior history of extrathoracic malignancy can be established on clinical grounds or by immunohistochemistry in some cases. The presence of multiple pulmonary nodules is usually considered as evidence of metastatic disease. However, in patients who present with a solitary lung nodule, particularly if they have history of squamous cell carcinoma, this distinction may be extremely difficult. Histologic criteria that are used by surgical pathologists include the comparison of histologic grade and identification of premalignant changes in the respiratory epithelium. The accuracy of this approach is uncertain. The correct diagnosis has practical importance for choice of therapy. The surgical approach and adjuvant therapy are usually different in these situations. Furthermore, patients with early-stage non-small cell lung carcinoma usually have better prognosis than patients with metastatic carcinoma. Several molecular assays can be used including PCR-based DNA clonality assays, mutational analysis, SNPs, and gene expression assays to distinguish between these two possibilities.
DNA-Clonality Assays
The clonal origin of a tissue may be assessed by analysis of somatic genetic alterations (e.g., loss of heterozygosity [LOH]) or chromosome X inactivation pattern (X-linked clonality analysis).1 These PCR clonality assays are based on microsatellite analysis. Microsatellites represent a highly polymorphic and repetitive noncoding DNA sequences widely distributed in the human genome. Several microsatellites have been used for DNA fingerprinting and are very useful in genetic linkage analysis. If the selected polymorphic region is related to known tumor suppressor gene, a possibility of mutation or function dysregulation of the corresponding gene can be determined based on observed alterations of microsatellite markers. Therefore, the assumption is that the LOH of a given genetic marker is linked to loss of tumor suppressor genes. Comparison of patterns of LOH between two tumors can help to determine whether the tumors are clonally similar (metastases) or different (independent primaries) (Fig. 21-1).2,3 and 4 There are several issues that should be
addressed.5 First, LOH assay requires matched normal DNA in order to determine whether an individual is heterozygous (informative) or homozygous (noninformative) for a particular chromosomal locus. Normal DNA may be obtained from morphologically normal-appearing tissue or from the patient’s leukocytes, which may not be readily available. The second issue is reliability of the results. Some authors report that LOH assay can definitely establish whether tumor represents a metastasis or a new primary lung cancer in 91% of cases.3 In our experience, a number of selected microsatellite markers and selection of chromosomal loci may influence the results and interpretation. Other considerations include tumor cell heterogeneity, sample size, tissue control, artifactual allelic dropout, cost, and turnaround time.
addressed.5 First, LOH assay requires matched normal DNA in order to determine whether an individual is heterozygous (informative) or homozygous (noninformative) for a particular chromosomal locus. Normal DNA may be obtained from morphologically normal-appearing tissue or from the patient’s leukocytes, which may not be readily available. The second issue is reliability of the results. Some authors report that LOH assay can definitely establish whether tumor represents a metastasis or a new primary lung cancer in 91% of cases.3 In our experience, a number of selected microsatellite markers and selection of chromosomal loci may influence the results and interpretation. Other considerations include tumor cell heterogeneity, sample size, tissue control, artifactual allelic dropout, cost, and turnaround time.
X-linked clonality analysis can be used only for clonality analysis in informative females, does not assess tumor heterogeneity, and does not provide any data on the precise genetic alteration responsible for clonal proliferation. In females, each X chromosome may be inactivated through a random methylation process early in embryogenesis. Once established in a particular cell, the methylation pattern is invariably transmitted to all the progenies. Hence, a unique nonrandom pattern of methylation is expected in malignant tumors, since all cells are supposed to derive from the same precursor. DNA probes can detect polymorphisms at particular X-linked loci to allow distinctions between the maternal X chromosome and paternal X chromosomes in a female subject. The active X chromosome may be distinguished from the inactive X chromosome by its state of methylation or by gene expression. Ideally, half the cells in a polyclonal reactive tissue will have inactivated the paternally derived X chromosome and half will have inactivated the maternally derived X chromosome. A monoclonal or clonal population of cells by contrast will have exclusively inactivated one X chromosome. There are several polymorphic foci that have been used to assess clonality by this assay. The X-linked human androgen receptor gene (HUMARA gene) is particularly well suited for analysis because it has a high rate of heterozygosity (“informativeness” >90%)
and consistent patterns of methylation. While nonrandom patterns of X chromosome inactivation may occur in up to 25% of female subjects (skewed lyonization), assessment of the HUMARA assay can be helpful in assessing selective highly concentrated cellular elements. This assay is technically challenging and is less suitable for the clinical practice than LOH assay.
and consistent patterns of methylation. While nonrandom patterns of X chromosome inactivation may occur in up to 25% of female subjects (skewed lyonization), assessment of the HUMARA assay can be helpful in assessing selective highly concentrated cellular elements. This assay is technically challenging and is less suitable for the clinical practice than LOH assay.
Mutational Analysis
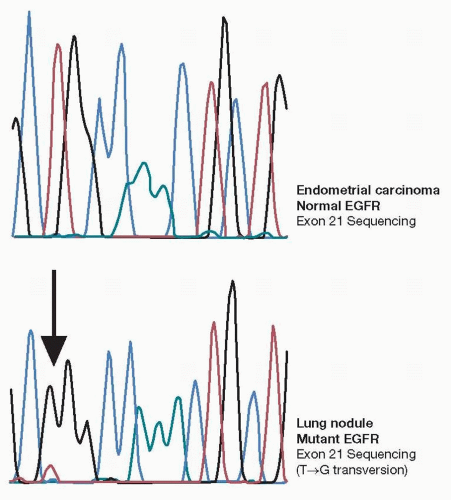
Several reports in literature indicate significance of DNA mutational analysis of p53 and other genes by DNA direct sequencing or other mutation detection techniques.6,7 Our recent experience indicates that detection of EGFR mutations may not only predict patients’ response to TKI therapies but also can be of diagnostic importance. Somatic mutations of EGFR unique to lung adenocarcinoma have been independently reported by several groups. Although several reports indicated the presence of these mutations in colorectal carcinomas and head and neck carcinomas, recent reports question those results. The differentiation of lung adenocarcinoma from endometrial carcinoma, breast, or pancreatic adenocarcinomas may be difficult based on morphology and immunohistochemistry. EGFR mutations in exons 18 to 21 may be extremely useful in making this distinction (Fig. 21-2). Unfortunately, these mutations occur in only 10% of Western population. On the other hand, up to 50% of Asian patients may harbor these mutations, and therefore, this assay may be more suitable for diagnostic use in this subset of patients.
ORIGIN OF TUMORS IN LUNG ALLOGRAFTS
The origins of malignancies that develop after transplantation of lung allografts are not always clear. In our experience, the most common malignancies in lung allografts are primary lung carcinomas. However, more challenging cases are metastatic carcinomas to the lung, which may represent either incidentally transmitted occult tumors from clinically healthy donors to the recipients or metastasis of a recipient’s primary tumor outside of the lung. To resolve the question of donor
versus recipient tumor origin, DNA typing can be performed.8,9 There are several commercially available diagnostic kits containing short tandem repeats (STRs). Population statistics for these genetic loci have been determined and are used in the calculation of genetic profile frequencies. DNA typing is performed as a multiplex PCR, which involves adding more than one set of PCR primers to the reaction in order to target multiple locations throughout the genome. This is an ideal technique for DNA typing because the probability of identical alleles in two individuals decreases with an increase in the number of polymorphic loci examined. Many commercial DNA typing kits also have the amelogenin gene, which is located on the X and Y chromosomes and is useful in determining the sex of an individual. The assay includes analysis and comparison of genomic DNA from the tumor in question and the recipient DNA (either from tissue or peripheral blood). If tumor and recipient DNA match, then the tumor is of recipient origin; if they are different, the tumor came from the donor (Fig. 21-3).
versus recipient tumor origin, DNA typing can be performed.8,9 There are several commercially available diagnostic kits containing short tandem repeats (STRs). Population statistics for these genetic loci have been determined and are used in the calculation of genetic profile frequencies. DNA typing is performed as a multiplex PCR, which involves adding more than one set of PCR primers to the reaction in order to target multiple locations throughout the genome. This is an ideal technique for DNA typing because the probability of identical alleles in two individuals decreases with an increase in the number of polymorphic loci examined. Many commercial DNA typing kits also have the amelogenin gene, which is located on the X and Y chromosomes and is useful in determining the sex of an individual. The assay includes analysis and comparison of genomic DNA from the tumor in question and the recipient DNA (either from tissue or peripheral blood). If tumor and recipient DNA match, then the tumor is of recipient origin; if they are different, the tumor came from the donor (Fig. 21-3).
MOLECULAR DIAGNOSIS OF BRONCHIAL MUCOEPIDERMOID CARCINOMA
The t(11;19)(q21;p13) is the major chromosomal abnormality observed in salivary gland mucoepidermoid carcinomas.10,11,12 and 13 This translocation involves two genes: mucoepidermoid carcinoma translocated 1 (MECT1) gene located on chromosomes 19p13 and a mammalian mastermind-like 2 (MAML2) located on 11q21.10,12,13 MECT1-MAML2 fuses exon 1 of MECT1 with exons 2 to 5 of MAML2 (Fig. 21-4). MECT1 is a coactivator of cyclic AMP responsive element binding protein. MAML2 is an essential coactivator of NOTCH receptor transcriptional activation and signaling. The translocation disrupts NOTCH signaling, activating transcription of NOTCH target genes, and stimulates aberrant activation of downstream cyclic AMP/cyclic AMP responsive element binding protein-signaling genes. Similar to head and neck mucoepidermoid
carcinomas, this translocation is exclusively seen in bronchopulmonary mucoepidermoid carcinomas when compared to primary lung squamous, adenosquamous, and adenocarcinomas.14
carcinomas, this translocation is exclusively seen in bronchopulmonary mucoepidermoid carcinomas when compared to primary lung squamous, adenosquamous, and adenocarcinomas.14
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree