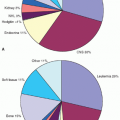
the number of individual proteins. There are powerful emerging technologies to measure global protein expression profiles, including protein arrays, mass spectroscopy, isotope-coded affinity tags (ICAT), isobaric tags for relative and absolute quantitation (iTRAQ), and stable isotope labeling by amino acids in cell culture (SILAC), that are able to generate quantitative global profiles of the proteome and phospho-proteome.12 Although application of molecular technologies for proteomic analysis of clinical samples have lagged behind genomics in terms of sensitivity, robustness, and reproducibility, these methods are likely to increase significantly in the future in their utility to biologically characterize cancer and provide clinically relevant biomarkers.
TABLE 3.1 Inherited Predisposition to Cancer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 Figure 3.2 Whole-genome and proteome investigation of cancer. The human genome is contained within 23 chromosomal pairs comprising 3.2 billion pairs of the four nucleotides (adenosine [A], cytidine [C], guanosine [G], and thymidine [T]). Two percent of the genome is transcribed into 20 to 25,000 protein-coding messenger RNAs (mRNA). The genomic sequence contains a promoter region, exons (containing the coding regions), and introns. The introns are spliced out following transcription, and alternate splicing can generate several different mRNAs and protein products. Many regions of the genome are also transcribed into noncoding RNA molecules (>10,000), including microRNAs (˜700). Shown on the right are some of the methodologies in genomics, including low-resolution chromosomal structure analysis such as cytogenetics and molecular cytogenetics (fluorescent in-situ hybridization [FISH], comparative genomic hybridization [CGH], and spectral karyotyping [SKY]). Genetic mapping uses DNA markers to find linkage of a genomic region to an inherited disease to eventually identify the causal gene. Physical mapping uses clones containing human nucleic acids (e.g., bacterial artificial chromosome) to define physical locations of genes and markers within each chromosome. In parallel to the sequencing of the human genome, many single nucleotide variations (SNVs) or single nucleotide polymorphisms (SNPs) are being detected and cataloged and may contribute to the phenotypic differences found in many patients and their cancers. Increasingly, “next generation” sequencing methods for profiling and discovery of novel genes are replacing traditional genomics methods. Finally, several methods are available for detecting which genes are actively being transcribed and translated into proteins, including protein arrays, mass spectroscopy, and isotope-coded affinity tags (ICAT).225 (Figure modified from references.225,226) |
DNA onto a metaphase preparation of normal human chromosomes. However, this approach has been progressively replaced by higher resolution methods including array-based comparative genomic hybridization (A-CGH), using arrays based on bacterial artificial chromosome (BAC), cDNA, oligonucleotides, SNPs, and, most recently, NGS methods.
fusion gene in an area of extensive DNA chromosomal rearrangement in rhabdomyosarcoma.48 Such discoveries will not be discovered by traditional cytogenetic techniques or DNA sequencing; therefore, RNAseq is an effective method to identify gene fusions, including novel or rare events.
 Figure 3.4 The types of genomic alterations that can be detected by next-generation sequencing methods. The flexibility of next-generation sequencing allows for the detection of multiple different genomic alterations. DNA or cDNA generated from RNA is fragmented, and sequence reads are produced. The reads are composed of sequenced regions (depicted here as the colored bars) and spacer regions (depicted here in grey). These generated short sequences are aligned to a reference genome in this case; the reads align to chromosome 2 and 13. From the alignment of many short reads, a consensus sequence emerges in which multiple genomic variations can be detected including point mutations (Reference A changes to a C in this example); small insertions and deletions; deletions including homozygous regions (no generated reads) and heterozygous regions (only half the expected reads are generated); amplifications (more reads are generated); and translocations where the generated sequences align to different genomic locations (in this case, chromosome 2 and 13). (Figure adapted from Meyerson 2010.227) |
de novo germline mutation) is estimated to comprise about 40% of affected persons. These patients often have early onset, bilateral disease, with a positive family history for retinoblastoma, leading Knudson to propose a “two-hit” mechanism of carcinogenesis in which the first genetic defect, already present in the germline, must be complemented by an additional spontaneous mutation before a tumor can arise. In contrast, the sporadic form results when two spontaneous mutations take place in the same cell.57
receptor (either an IG or TCR) gene. The proto-oncogene present on the translocated chromosome then becomes activated via the regulatory region of the antigen receptor gene. The notion that these translocations are the result of illegitimate V(D)J recombinase activity is strengthened by the presence of features associated with normal V(D)J recombinase action, such as site-specific DNA cleavage at cryptic heptamer sequences and the addition of nontemplated (“N” region) nucleotides at the translocation breakpoints.64 Interestingly, there are now a number of examples of fusions between nonantigen receptor genes that show all of the aforementioned hallmarks of normal V(D)J recombinase activity.65
TABLE 3.2 Recurrent Chromosomal Translocations Associated with Hematologic Malignancies | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
histiocytic lymphoma and has been shown to differentiate along the macrophage lineage in vitro. Subsequently, this fusion was found in patients with a wide spectrum of hematologic malignancies, but most commonly in patients with T-cell ALL.79 CALM-AF10 fusions were identified in 12 (9%) of 131 consecutive patients with T-cell ALL; all patients with CALM-AF10 fusions had either immature T-cell lymphoblasts that expressed no TCR genes or TCRγ/δ-positive lymphoblasts.80
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree