Chapter Outline
PRENATAL ORIGIN OF INFANT LEUKEMIA
BIOLOGY OF MIXED-LINEAGE LEUKEMIA TRANSLOCATIONS
FUTURE DIRECTIONS IN INFANT ACUTE LEUKEMIA TREATMENT
The biologic features and clinical characteristics of infant leukemias differ significantly from those of leukemias in older children. Infant leukemias are distinguished not only by the young age of patients at diagnosis but by their unique morphologic, immunologic, clinical, and genetic presentation. For example, acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) present in infants with a somewhat unique constellation of clinical features, including hyperleukocytosis, massive organomegaly, and central nervous system (CNS) involvement at diagnosis. Biologically, the most notable feature distinguishing infant leukemia from leukemia in older children is the high incidence of rearrangements involving the mixed-lineage leukemia (MLL) gene * located on chromosome band 11q23. In the case of infant ALL, unique clinical and biologic characteristics are accompanied by an exceptionally poor prognosis, which is in stark contrast to the cure rates achieved for older children with ALL. In this chapter, the epidemiologic, biologic, and clinical features and the treatment of infant ALL and AML will be discussed.
* In this chapter, KMT2A will be referred to as MLL. ALL1 and HRX are also alternative names for this gene.
Epidemiology
The annual incidence of leukemia in the first year of life in the United States is approximately 40 cases per million children. Infants account for 2.5% to 5% of ALL cases and 6% to 14% of AML cases in childhood. Unlike in older children with leukemia, in whom the percentage of ALL cases is approximately four times that of AML, in infants the ratio of ALL to AML is approximately equal. Furthermore, in contrast to an excess of males among older children with leukemia, a slight female predominance is seen in infants with this disease. Although neuroblastoma represents the most common neoplasm in infants, leukemia is the leading cause of death caused by neoplastic disease in this age group ( Table 52-1 ).
| Histology | Incidence (per million) |
|---|---|
| Neuroblastoma | 64.6 |
| Leukemias | 40.5 |
| Central nervous system | 29.7 |
| Retinoblastoma | 26.7 |
| Wilms tumor | 22.5 |
| Germ cell | 15.3 |
| Soft tissue | 15.2 |
| Hepatic | 9.5 |
| Lymphomas | 4.4 |
| Epithelial | 2.8 |
| Other, unspecified | 1.2 |
| Bone | 0.5 |
Prenatal Origin of Infant Leukemia
The onset of leukemia in infancy strongly suggests a prenatal leukemogenic event, and significant molecular and epidemiologic evidence supports this hypothesis. Molecular studies of monozygotic twins with concordant ALL have demonstrated identical clonal, nonconstitutional rearrangements of the MLL gene and identical oligoclonal, heavy-chain immunoglobulin gene rearrangements in the peripheral blood of these infants. This finding suggests the occurrence of a single in utero leukemogenic event in one twin with generation of a clone that is passed to the other fetus by intraplacental metastasis. Further evidence of oncogenesis in utero comes from reports of fetal deaths caused by AML with an MLL gene rearrangement. Moreover, identical MLL gene fusions have been traced back to neonatal Guthrie genetic screening cards of children who were diagnosed with ALL in their first 2 years of life. Similarly, leukemia has been diagnosed in fetuses with Down syndrome as early as 33 weeks’ gestation, and the characteristic GATA binding protein 1 (GATA1) gene mutation associated with megakaryocytic leukemia in young children with Down syndrome has been identified in the neonatal Guthrie genetic screening cards of children in whom leukemia later developed.
Risk Factors
As with most cancers, the causes of infant leukemia remain largely unknown. However, several epidemiologic risk factors associated with an increased risk of leukemia in young children have been identified. Given the close temporal relationship between embryogenesis and the clinical diagnosis of cancer, most investigations have focused on maternal characteristics and in utero exposures. Maternal alcohol consumption during pregnancy has been correlated with an increased risk of infant AML. Studies have also shown an increased incidence of infant leukemia, particularly ALL, in infants with birth weights higher than 4000 g. It has been speculated that high levels of endogenous insulin-like growth factors associated with high birth weight may contribute to leukemogenesis. Other proposed risk factors include maternal pesticide and solvent exposure, in utero radiation exposure, and an adverse maternal reproductive history. However, given the rarity of infant leukemia, it has been difficult to connect the development of leukemia with any of these exposures or clinical features definitively.
Of particular interest is the potential relationship between maternal consumption of naturally occurring deoxyribonucleic acid (DNA) topoisomerase II inhibitors and the development of infant leukemia. MLL gene rearrangements, similar to those found in many infant leukemias, are common in secondary acute leukemias arising after exposure to DNA topoisomerase II inhibitors, including the epipodophyllotoxins (e.g., etoposide). This relationship has led to the hypothesis that transplacental exposure to naturally occurring topoisomerase II inhibitors may be involved in the pathogenesis of infant leukemia. This hypothesis is supported by the finding that both primary infant MLL gene–rearranged leukemias and therapy-related secondary acute leukemias have MLL gene break points that are similarly distributed within the MLL gene break point cluster region (BCR). Furthermore, several dietary bioflavonoids, such as quercetin (found in certain fruits and vegetables) and genistein (found in soybeans) are known topoisomerase II inhibitors and have been shown to induce MLL gene cleavage in vitro. In a study by the Children’s Oncology Group (COG), it was found that increased maternal consumption of these and other naturally occurring topoisomerase II inhibitors such as catechins (found in red wine, tea, and cocoa) is associated with an increased risk of MLL gene–rearranged infant AML but not infant ALL. This association, if true, likely only accounts for a small proportion of infant leukemia cases. Furthermore, the fact that many of these foods are commonly consumed and infant leukemia is rare suggests that this association is either weak or requires a combination of factors. Thus one cannot make recommendations to restrict certain foods to decrease the likelihood of the development of leukemia. Moreover maternal consumption of fruits and vegetables during pregnancy has been associated with a decrease rather than an increase in the risk of infant leukemia overall.
Pharmacogenetic differences in the metabolism of topoisomerase II inhibitors have been hypothesized to modulate the relationship between exposure to these chemicals and the occurrence of MLL gene rearrangements. A common structural feature shared by many topoisomerase II inhibitors, including bioflavonoids, is a quinone moiety. Quinone metabolites generated as by-products of these compounds in the fetal liver have been shown to cleave the MLL gene. Quinones are normally detoxified by reduced nicotinamide adenine dinucleotide phosphatase quinone oxidoreductase (NQO1). The presence of a low-activity variant of NQO1 has been associated with an increased risk of MLL gene–rearranged infant ALL. However, a similar study has also demonstrated a relationship between this NQO1 polymorphism and infant ALL without MLL gene rearrangements.
Biology of Mixed-Lineage Leukemia Translocations
The MLL gene located on chromosome band 11q23 is commonly altered in infant leukemia in that it is rearranged in 80% of cases of infant ALL and 60% of cases of infant AML. These chromosomal abnormalities usually involve reciprocal translocations, which encode chimeric transcripts that give rise to oncogenic fusion proteins with pronounced transforming potential. A number of groups cloned the MLL-AF4 (AFF1) gene in the early 1990s. The MLL-AF4 gene encodes a protein of 2304 amino acids, with the NH2-terminal 1439 amino acids derived from the MLL gene on chromosome 11 and COOH-terminal 865 amino acids derived from the AF4 gene on chromosome 4. Subsequently, close to 100 different translocations have been identified, all of which appear to produce a fusion protein possessing the NH2-terminus of MLL fused in-frame to the COOH-terminus of the fusion partner ( Fig. 52-1 ).
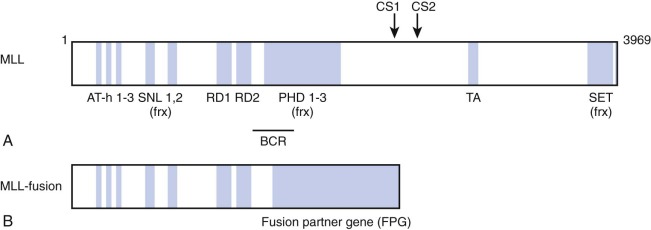
The MLL gene encodes a 3969–amino acid DNA-binding protein that possesses multiple recognizable protein motifs, including an NH2-terminal DNA binding domain, transcriptional activation and repression domains, and a COOH-terminal Su(var)3-9, Enhancer of Zeste, Trithorax (SET) domain that contains histone methyltransferase activity. Biochemical studies have identified MLL as a member of a large multiprotein complex that contains members involved in chromatin modification and remodeling. Notably, the complex includes histone deacetylases and members of the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex. Also MLL is recruited to the promoters of select cell cycle regulatory genes by the protein product of the multiple endocrine neoplasia 1 (MEN1) tumor suppressor gene, suggesting a role for MLL in tumor suppression and cell cycle control. These data support the hypothesis that the MLL protein regulates gene expression via chromatin modification.
Analysis of Mll knockout mice has suggested that Mll plays an important role in development and hematopoiesis through maintenance of appropriate homeotic (Hox) gene expression. Detailed studies assessing the specific role of Mll in hematopoietic development have shown that Mll is necessary for definitive hematopoiesis and for the expansion of hematopoietic progenitors and stem cells found in the aorta-gonad-mesonephros region of the developing embryo. The defect in hematopoietic progenitor expansion can be rescued by reexpression of Hox genes, confirming the importance of Mll-mediated Hox gene expression during hematopoiesis.
Multiple studies have demonstrated the ability of Hox genes to induce leukemia in mice, and the t(7;11)(p15;p15) translocation found in some human acute myeloid leukemias results in a fusion of the homeobox A9 (HOXA9) gene to the nucleoporin 98kDa (NUP98). Given the apparent importance of HOX genes in leukemogenesis, it seems likely that translocations involving MLL, a known regulator of HOX genes, alters expression of HOX genes that are important for leukemogenesis. Further support for HOX genes as central regulators of MLL gene–induced leukemogenesis comes from gene expression studies that have found multiple HOXA cluster genes more highly expressed in MLL gene–rearranged myelogenous and lymphoblastic leukemias compared with MLL gene germline leukemias.
Gene expression studies of human MLL gene–rearranged B-precursor ALL have demonstrated that hundreds of genes are differentially expressed when compared with other B-precursor ALLs. Based on the magnitude of the differences in gene expression, it appears that MLL gene translocations specify a unique lymphoblastic leukemia. Other large gene expression studies have also shown that ALLs with distinct chromosomal rearrangements have unique gene expression profiles, providing support for this hypothesis. The genes that are relatively highly expressed in MLL gene–rearranged B-precursor ALL are those associated with hematopoietic progenitors and developing myeloid cells, whereas the genes expressed at lower levels are genes associated with lymphoid identity. Studies have also defined specific gene expression signatures associated with MLL gene translocations in pediatric—and in some cases infant—AML blasts. It is of interest that even though clear differences exist in expression of lineage-associated genes between MLL gene–rearranged ALL and MLL gene–rearranged AML, a core gene expression profile appears to be found in all MLL gene–rearranged human leukemias, independent of the lineage markers. Presumably, MLL fusion proteins directly regulate a subset of these genes. This theory is further supported by the fact that this MLL-associated signature consists of multiple highly expressed HOX genes.
The observation that leukemic cells bearing an MLL gene translocation often coexpress both myeloid and lymphoid markers raises the possibility that MLL fusion genes selectively transform hematopoietic stem cells (HSCs). If so, this HSC population would have an inherent self-renewal capacity that could be co-opted for leukemogenesis. Xenograft transplantation studies have demonstrated that a rare subpopulation of CD34+ and CD38− human myeloid leukemia cells are able to transfer the disease to immunodeficient mice, providing support for HSCs as the normal compartment from which leukemia-initiating cells (leukemia stem cells) might arise. Alternatively, the translocation event between the MLL gene and a partner gene may affect a more committed population. In this scenario, the MLL fusion protein might confer a self-renewal and proliferative capacity to these short-lived progenitors and allow them to initiate leukemogenesis. Studies have suggested that Mll fusion proteins and potentially other translocation-associated fusion proteins are capable of inducing leukemia when expressed in fully committed myeloid progenitors, such as granulocyte-macrophage progenitors. These findings are of particular importance because they suggest that products of chromosomal translocations found in human leukemias are able to induce a program of self-renewal that is not normally present in hematopoietic progenitors.
Multiple lines of evidence now point to a multistep pathogenesis of human acute leukemia. Elegant epidemiologic studies have suggested that childhood leukemias require at least two and probably more genetic events to occur for the development of leukemia. In particular, lymphoblastic leukemias with ETV6-RUNX1 (TEL-AML1) rearrangements appear to develop after a multistep process. TEL-AML1 rearrangements have been detected in blood taken at birth from a child in whom ALL then developed 3 to 5 years later. This finding suggests that TEL-AML1 rearrangements are the first genetic event but that other mutations also are required for the development of ALL. Similar studies have been performed on blood spots from children in whom MLL gene–rearranged ALL subsequently developed, and the MLL gene translocations clearly develop in utero, even though the leukemias become apparent at some point during the first year of life.
Receptor tyrosine kinases are attractive candidates as signaling molecules that may cooperate with translocation-associated fusion proteins during leukemogenesis. Ever-increasing evidence has suggested that activated kinases play a central role in the pathogenesis of leukemias and myeloproliferative syndromes. The most dramatic evidence for such a role is activation of the Abelson (ABL) tyrosine kinase by the BCR-ABL fusion produced by t(9;22) and its inhibition by imatinib (Gleevec). Other mutant kinases frequently identified in AML and subsets of ALL are the receptor tyrosine kinases fms-related tyrosine kinase–3 (FLT3), c-KIT, and platelet-derived growth factor receptor, among others. Given the high level expression and recurrent mutation of FLT3 in MLL gene–rearranged ALL, studies are under way to assess FLT3 inhibitors in patients with MLL gene–rearranged ALL (discussed later). As approaches to target the rat sarcoma (RAS) signaling pathway are further developed, RAS inhibitors might also be considered in RAS-mutant cases.
Given the central importance of the MLL fusion protein generated by the MLL gene translocation, significant effort has been directed toward defining a unifying mechanism of oncogenesis, because it would facilitate pharmacologic targeting of shared leukemogenic mechanisms. Some broad patterns have emerged that have focused on mechanisms that control MLL target gene expression. The most commonly occurring MLL gene translocations generate chimeric fusion proteins that harbor the NH3-terminus of MLL fused to proteins that are normally part of nuclear complexes, the function for which is now emerging. Nuclear proteins such as AF4, AF9, ENL, ELL, AF10, AF17, and AFF4—fusions of which together account for the vast majority of MLL leukemias—normally directly or indirectly associate with complexes that regulate gene expression. A number of complexes linked to transcriptional elongation have been reported, often with overlapping protein components, such as the ENL-associated protein (EAP) complex, the AF4/ENL/P-TEFb (AEP) complex, the super elongation complex (SEC), and the complex comprising the disruptor of telomeric silencing (DOT1)–like histone 3 lysine 79 (H3K79) methyltransferase (DOT1L; DOT1L containing complex [DotCom]; Fig. 52-1 ). These data point to aberrant control of transcriptional elongation as critical for MLL fusion–mediated oncogenesis.
The histone methyltransferase DOT1 is a non-SET domain containing histone methyltransferase solely responsible for catalyzing the methylation of H3K79. H3K79 methylation is associated with most genes that are highly expressed in hematopoietic cells and is believed to be involved either in activation or maintenance of gene expression. Thus a prominent hypothesis is that MLL fusion proteins induce aberrant gene expression via recruitment of DOT1L (among other protein complexes) to MLL target genes, including the HOXA cluster genes. Genome-wide studies have demonstrated elevated H3K79 methylation at MLL target genes in MLL gene–rearranged ALL and AML cells. Several recent studies using conditional loss of function mouse models and ribonucleic acid (RNA) interference approaches have formally demonstrated a critical role for DOT1L in MLL fusion–driven leukemias. These studies demonstrate that genetic inactivation of DOT1 leads to a decrease in MLL fusion target gene expression, including a rapid decrease in HOXA cluster gene expression that is correlated with an antiproliferative response. Remarkably, inactivation of DOT1L does not affect the transformation potential of a number of other leukemogenic oncogenes. Furthermore, microarray-based gene expression studies showed that MLL-fusion target gene expression is much more dependent on DOT1L than is gene expression broadly. These studies highlight the importance of aberrant H3K79 methylation for the transforming activity of MLL fusion proteins including MLL-AF4, MLL-AF9, MLL-AF10, and MLL-ENL and show that DOT1L is required for continued HOXA cluster gene expression. These results potentially have profound clinical implications because these fusions comprise the vast majority of MLL gene–rearranged leukemias; small-molecule DOT1L inhibitors have been developed and are entering clinical trials (discussed later).
Acute Lymphoblastic Leukemia
Clinical and Biologic Features
ALL in infants is characterized by a high leukocyte count at presentation, marked hepatosplenomegaly, and a relatively high incidence of CNS involvement. The immunophenotype of infant ALL is usually that of an immature B-lineage precursor and is characterized by a lack of cluster of differentiation (CD)–10 expression, as well as by the coexpression of myeloid-associated antigens, such as CD14, CD15, and CDw65. Lack of CD10 expression and coexpression of myeloid markers correlates with the presence of an MLL gene rearrangement, which is present in 80% of infants who have ALL compared with only 2% to 4% of older children who have ALL. MLL gene rearrangements are more frequent in younger infants, with approximately 90% of infants younger than 6 months at diagnosis having MLL gene rearrangements in their leukemic blasts compared with 30% to 60% of infants aged 6 to 12 months. The most common MLL gene translocation observed in infant ALL is t(4;11)(q21;q23), resulting in the MLL-AF4 fusion. This translocation is found in approximately 70% of MLL gene–rearranged infant ALL cases. Other common translocations include t(11;19), resulting in the MLL-ENL fusion, and t(9;11), resulting in the MLL-AF9 fusion, which occur in 15% and 4% of MLL gene–rearranged infant ALL cases, respectively. Cases of infant ALL without an MLL gene rearrangement generally resemble the more common B-precursor ALL phenotype (CD19+, CD10+) typical of older children with ALL. Chromosomal abnormalities known to be favorable in older children with ALL, such as high hyperdiploidy (51 to 65 chromosomes, or a DNA index more than 1.16) and the TEL-AML1 fusion gene, are notably absent in infants. The distinguishing presenting characteristics of infants with ALL are summarized in Table 52-2 .
| Characteristic | Age (yr) | |
|---|---|---|
| 0-1 (%) | 1-18 (%) | |
| WBC * >100,000 | 58 | 6.3 |
| CNS-positive * | 14 | 1.5 |
| Phenotype | ||
| B lineage | 96 | 86.5 |
| T lineage | 4 | 13.5 |
| CD10-negative | 54.7 | 3.3 |
| Myeloid antigen coexpression | 28 | 5 |
| DNA index * >1.16 | 1.5 | 24.7 |
| TEL-AML1 * rearrangement | 4.5 | 24.1 |
| MLL rearrangement | 70-80 | 2-4 |
A striking feature of MLL gene–rearranged infant ALL is the paucity of secondary genomic alterations compared with older children who have pre–B-ALL. A mutational profiling study of oncogenic RAS alterations in MLL gene–rearranged ALL samples identified mutations in 15 of 109 samples (14%). Of the 38 samples with a t(4;11) MLL gene rearrangement, 9 (24%) were found to have RAS mutations. Identification of a RAS mutation was found to be an independent poor prognostic risk factor.
Prognostic Factors and Outcomes
Despite extraordinary improvements in the cure rates for older children with ALL, the prognosis for infants with this disease remains poor. Historically, infants with ALL who were treated with standard regimens had a less than 20% event-free survival (EFS). During the past decade, intensified regimens designed especially for infants have resulted in improved rates of EFS ranging from 28% to 54% ( Table 52-3 ). In contrast, EFS rates are greater than 80% for older children with ALL. Although 90% to 95% of infants with ALL achieve remission after initial induction chemotherapy, a favorable outcome is hampered by an exceedingly high relapse rate, typically within the first year after diagnosis.
| Study | Publication Date | Patients Enrolled | CR Rate (%) | EFS Time Point (yr) | EFS (%) |
|---|---|---|---|---|---|
| DFCI (1985-1995) | 1997 | 23 | 96 | 4 | 54 |
| Interfant-99 | 2007 | 482 | 94 | 5 | 45 |
| AIEOP-91, 95 | 2006 | 52 | 96 | 5 | 45 |
| BFM 86, 90, 95 | 1999 | 129 | 95 | 8 | 43 |
| CCG-1953 | 2006 | 115 | 97 | 5 | 42 |
| P9407 | 2012 | 97 | n/a | 5 | 41 |
| CCG-1883 | 1999 | 135 | 97 | 4 | 39 |
| CCG-107 | 1999 | 99 | 94 | 4 | 33 |
| UKALL-92 | 2002 | 86 | 94 | 5 | 33 |
| POG 8493 | 1997 | 82 | 93 | 4 | 28 |
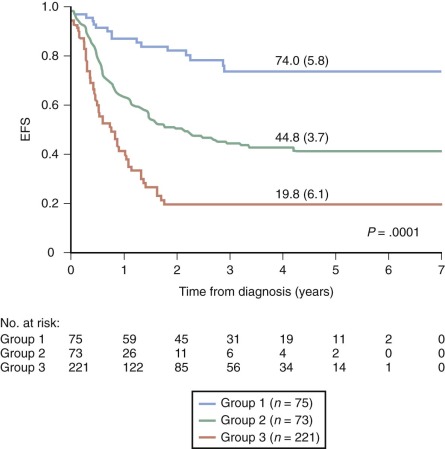
Several closely related adverse prognostic factors have been identified for infants with ALL, including the presence of an MLL gene rearrangement, lack of CD10 expression, coexpression of myeloid markers, age less than 6 months at diagnosis, high white blood cell (WBC) count at presentation, CNS involvement, and poor early response to prednisone therapy. In multivariate analyses, the presence of an MLL gene rearrangement, age younger than 6 months, and a poor early response to prednisone therapy consistently emerge as the most important adverse prognostic features. In many studies, the presence of an MLL gene rearrangement is the most important independent predictor of outcome for infants. Long-term EFS for infants with MLL gene–rearranged ALL has ranged from 13% to 34%, versus 52% to 95% for infants with germline MLL ( Fig. 52-2 ).
In addition to MLL gene status and age, early response to therapy has been shown to be an important predictor of outcome in infants with ALL. The Berlin-Frankfurt-Munster group has identified a poor response to prednisone as one of the strongest predictors of outcome for infants with ALL, regardless of MLL gene status. Infants with ALL who have a poor response to a 7-day prednisone monotherapy prophase (defined as more than 1000 blasts/µL present in peripheral blood on day 8) had a 6-year EFS of only 15%, compared with a 6-year EFS of 53% for infants with a good response to prednisone. This finding was reproduced in the most recent clinical trial conducted by the Interfant study group (Interfant 99), in which infants with a poor response to steroids had a 30% 4-year EFS, compared with a 56% 4-year EFS for good responders. Similarly, the Children’s Cancer Group found a threefold excess risk of treatment failure in infants whose bone marrows contained more than 5% leukemic blast cells after 14 days of multiagent chemotherapy.
It has been suggested that the specific MLL gene fusion partner might also have prognostic significance for infants with ALL. However, in a large retrospective study that included data from more than 200 cases of MLL gene–rearranged ALL in infants, no significant difference in outcome was found among cytogenetic subgroups, including t(4;11), t(9;11), t(11;19), and other 11q23 rearrangements. The Interfant study group also failed to find any association between the specific MLL gene translocations and treatment outcome. More recently, localization of the fusion break point has been identified as a potential prognostic biomarker, with break points within MLL intron 11 demonstrating poorer outcomes. Gene expression profiling has also identified candidate prognostic markers. Analysis of 97 samples from P9407, a COG trial for infant ALL conducted between 1996 and 2006, identified patients with low FLT3 expression as having superior outcomes, which had previously been observed in patients enrolled in the Interfant 99 trial. Patients with high FLT3 expression, in addition to either high iroquois homeobox 2 (IRX2) or high transforming, acidic coiled-coil containing protein 2 (TACC2) expression, had very poor 4-year EFS rates.
Treatment
Attempts to improve clinical outcomes for infants with ALL have generally involved intensifying treatment regimens and incorporating chemotherapeutic agents more commonly used in AML therapy. An ongoing international trial, Interfant 06, is evaluating the outcome of patients randomly treated with either a standard consolidation approach using mercaptopurine, cyclophosphamide, and cytarabine or two consecutive AML-based cycles that include cytarabine, etoposide, and both daunorubicin and mitoxantrone. Several preclinical and clinical findings have also informed the development of treatment regimens for infants with ALL, including clinical response to prednisone therapy and in vitro drug sensitivity profiles of infant ALL lymphoblasts. When compared with cells from older children with ALL, leukemic blasts from patients with infant ALL are more often resistant to glucocorticoids (prednisone and dexamethasone) and L-asparaginase in in vitro assays. Importantly, both of these agents are central components of current ALL therapeutic regimens. In vitro studies of sarcoma (Src) kinase inhibition demonstrated sensitization of MLL gene–rearranged ALL cell lines to glucocorticoids, highlighting this strategy as a possible future therapeutic option in these patients. Given the prognostic significance of prednisone response in ALL, some infant ALL treatment groups now risk-stratify infants on the basis of prednisone response and other common prognostic factors, including MLL gene status, age, and diagnostic WBC count.
Although infant ALL blasts are relatively resistant to some common ALL therapeutic agents, they are remarkably sensitive to cytosine arabinoside (ara-C; cytarabine), a drug commonly used in the treatment of AML. This increased sensitivity may be related in part to elevated expression of the human equilibrative nucleoside transporter–1, which allows cytarabine to permeate the cell membrane at low to moderate concentrations. Clinical data are accumulating to suggest that increased use of cytarabine may benefit infants with ALL. Investigators from the Dana-Farber Cancer Institute ALL Consortium first reported an improved outcome in a small number of infants treated with intensified therapy that included high-dose cytarabine. Subsequently the cooperative groups COG and Interfant incorporated high-dose cytarabine into their intensified therapeutic regimens, a strategy that is likely to have contributed to the improvements in EFS for infants with ALL observed during the past decade.
Hematopoietic Stem Cell Transplantation
The role of hematopoietic stem cell transplantation (HSCT) in first remission for infants with ALL is controversial. Small uncontrolled studies have suggested that HSCT may benefit infants with MLL gene rearrangements. Although encouraging, these results must be interpreted with caution, given the small sample sizes, the failure to control for waiting time from diagnosis to transplantation, and the use of total body irradiation in cytoreduction, which many groups are avoiding given the significant morbidity associated with radiation in this age group. The most recent Interfant study group trial (Interfant 99) identified a subgroup of high-risk patients who may benefit from HSCT, including those diagnosed before 6 months of age and who have either a poor steroid response or a high WBC count at diagnosis. In contrast, a larger retrospective analysis did not confirm a benefit of HCST for infants with ALL. Similarly, the COG found no benefit of transplantation compared with intensive chemotherapy alone. Further studies are needed to adequately define the role for stem cell transplantation in infants with ALL.
Toxicity and CNS-Directed Therapy
A major consideration in the treatment of ALL in infants is the significant potential for short- and long-term toxicity, given the intensity of treatment regimens and the lack of pharmacokinetic and pharmacodynamic studies to ensure optimal dosing of chemotherapeutic agents in infants. Of particular concern is the risk of debilitating neuropsychological sequelae, especially in infants receiving cranial radiation. Severe neurologic deficits and learning disabilities have been reported in long-term survivors of infant ALL who received cranial radiation. In almost all subsequent clinical trials, attempts have been made to reduce neuropsychological complications by minimizing, delaying, or eliminating cranial radiation. Currently most investigators favor eliminating radiation in infants with ALL, even those with CNS leukemia at diagnosis, relying instead on intensive systemic and intrathecal chemotherapy. Several observations have supported this approach, including outcomes from large cooperative group studies documenting very low CNS relapse rates (3% to 9%) with intensive systemic and intrathecal chemotherapy alone.
Acute Myeloid Leukemia
Clinical and Prognostic Features
Infant AML is characterized by a high incidence of myelomonoblastic (French-American-British [FAB] M4) or monoblastic (FAB M5) morphologic features, frequent CNS involvement, extramedullary disease (including skin involvement), and relatively high leukocyte counts at diagnosis.
MLL gene rearrangements are found in about 60% of infant AML cases, with the most common translocation being t(9;11), resulting in the MLL-AF9 fusion, followed by t(11;19), resulting in the MLL-ENL fusion. MLL gene rearrangements in infant AML are associated with a FAB M4 or M5 morphology and hyperleukocytosis. Other cytogenetic abnormalities in infant AML of the FAB M4 or M5 morphologic subtype include inv(16) and monosomy 7. Translocations commonly seen in older children with AML, such as t(8;21) and t(15;17), are rare in infants with AML.
Unlike in infant ALL, the prognostic factors that predict outcome for infant AML are not clearly defined. Several studies have identified a variety of potential prognostic factors for infants with AML, including presenting WBC count, FAB M4 or M5 morphology, gender, and the presence of an MLL gene rearrangement, but the findings of these studies have varied widely and are often contradictory. The difficulty in clearly identifying consistent prognostic factors in infant AML may point to more biologic heterogeneity in infant AML (compared with infant ALL), as well as significant variations in the definition of study groups and treatment regimens.
The prognostic significance of MLL gene rearrangements in infant AML is unclear. Several studies have found that MLL gene rearrangements lack prognostic significance in childhood AML, whereas at least one study has found a trend toward a worse outcome. Alternatively, several groups have demonstrated that the presence of a specific translocation, t(9;11)(p22;q22) or MLL-AF9, confers a favorable prognosis. For example, the investigators at St. Jude Children’s Research Hospital found the presence of t(9;11) to be an important prognostic factor for patients treated in four consecutive pediatric AML trials. The 5-year EFS for infants whose leukemia cells carried t(9;11) was 70% versus 25% for those with other cytogenetic abnormalities, including other MLL gene rearrangements.
The Berlin-Frankfurt-Munster Group recently reported on the clinical characteristics of 62 pediatric patients with AML who had t(8;16)(p11;p13) translocations. The average age of diagnosis was very young (1.2 years), with common features of erythrophagocytosis (70%), leukemia cutis (58%), and disseminated intravascular coagulation (39%). Strikingly, seven patients diagnosed within the first month of life underwent spontaneous remission of their disease, three of whom remained in remission at the time of publication.
Outcomes and Treatment
Whereas age-associated treatment results are clearly evident in childhood ALL, with infants faring significantly worse, similar age-related differences in outcomes have not been routinely observed in AML. In large clinical trials in children with AML, EFS rates of 22% to 73% have been reported for infants, which do not differ significantly from outcomes achieved in older children ( Table 52-4 ). Given that treatment outcomes do not differ by age group, there has been no compelling clinical justification for the development of unique treatment strategies for infants with AML. Current therapeutic regimens for infants and older children focus on intensive remission induction and consolidation chemotherapy, with HSCT reserved for patients with matched sibling donors and those who have poor prognostic features. Although treatment regimens for infants with AML do not currently differ from those for older children, some potential differences have been proposed. The observation that t(9;11) may be a favorable prognostic factor in infants with AML has resulted in controversy with regard to whether bone marrow transplantation in first remission should be considered in this subgroup of patients. Another possible exception includes infants with megakaryoblastic leukemia who are harboring t(1;22)(p13;q13); these infants appear to have a particularly poor prognosis and may be candidates for more aggressive treatment or innovative experimental therapy.
| Study | Study Period | Patients Enrolled | EFS Time Point | EFS (%) |
|---|---|---|---|---|
| St. Jude (AML80-91) | 1980-1987 | 28 | 5 | 32 |
| BFM (AML 93) | 1993-1998 | 112 * | 5 | 41 |
| MRC (AML10, 12) | 1988-2002 | 151 | 5 | 58 |
| POG (8821) | 1988-1993 | 122 * | 5 | 22 |
| Nordic (NOPHO-93) | 1993-2001 | 57 * | 5 | 54 |
| CCG 2891 | 1989-1995 | 116 | 8 | 71 |
| Japan (ANLL91) | 1995-1998 | 35 | 3 | 72 |
| French (LAME 89, 91) | 1988-1998 | 42 | 5 | 37.3 |
| FHCRC | 1995-1998 | 35 * | 3 | 72 |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree