Fluorescence In Situ Hybridization
* Seattle Children’s Hospital, 4800 Sand Point Way NE, A-6901, PO Box 5371, Seattle, WA 98105-0371
Keywords
• Fluorescence in situ hybridization • FISH • Interphase FISH • Metaphase FISH
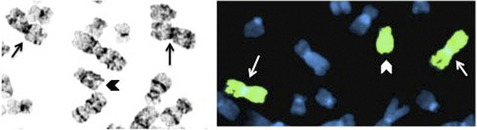
The basic elements of the FISH procedure include selection of probe(s) for sequence that is complementary to the target of interest, probe labeling, slide preparation, slide pretreatment, denaturation of probe and target, hybridization, washing, analysis, and interpretation. For genomic DNA analysis, FISH probes can target unique DNA sequences, repetitive DNA sequences, entire chromosome arms, or whole chromosomes. Unique sequence probes are useful for identifying gains and losses of specific genes or genomic regions, as well as genomic rearrangements, such as translocations. Repetitive sequence probes are primarily used for interrogating centromeric or pericentromeric regions. Probes that hybridize along the length of entire chromosome arms or whole chromosomes, also known as chromosome paints, can be used to confirm or further characterize chromosomal rearrangements identified by banded chromosome analysis (Fig. 1). Most FISH probes used for detecting unique sequences are currently made from bacterial artificial chromosomes that contain genomic inserts on the order of 100 to 300 Kb. Propagation and extraction of DNA from bacterial artificial chromosomes is relatively straightforward, and the size of the insert produces signal that can be easily visualized. Probes can also be made from vectors containing smaller inserts, such as fosmids or even plasmids, particularly if the target consists of repetitive DNA that will result in a larger signal. FISH using oligonucleotide probes was originally limited to repetitive targets; however, recent advances in the synthesis of long oligonucleotides promises to provide a source of probes that can be used to visualize nonrepetitive targets at a much greater resolution than can be achieved using large insert clones.1
Slide preparation, pretreatment, and other aspects of the FISH procedure vary depending on specific applications, and some of the relevant variables are discussed within the specific techniques or applications that follow. There are also unique issues related to signal analysis and interpretation depending on the specific FISH application, although recommendations that apply to many situations encountered in clinical laboratories are available.2 With the exception of FISH tests that are approved by the US Food and Drug Administration (FDA), clinical laboratories must validate FISH probes prior to use. For the detection of nonmosaic abnormalities, steps involved in this validation process include confirmation of correct probe localization, and assessment of probe sensitivity and specificity. For mosaic conditions or minimal residual disease detection, additional validation is required. Details on performing these validation steps are published elsewhere.2
FISH Techniques
Comparative Genomic Hybridization
Comparative genomic hybridization (CGH) is a method for detecting genomic gains and losses (copy number changes) that can be performed on either metaphase chromosome preparations or arrays. For metaphase-based CGH, a test DNA is labeled in one fluorophore, a reference DNA is labeled in another fluorophore, and equal amounts of labeled test and reference DNA are hybridized to metaphase chromosome preparations from a normal specimen. The ratio of fluorescence intensity of the hybridized test and reference DNA is measured along the length of the chromosomes using digital image analysis. Regions of loss in the test DNA result in a decreased ratio of test to reference DNA over those chromosome regions, whereas gains in the test DNA result in an increased ratio. Metaphase-based CGH requires only DNA from the test specimen, because the hybridization is performed on metaphase cells from a normal specimen. Thus, this technique was useful in the past for detection of copy number alterations in neoplastic specimens that were highly complex and not readily amenable to G-banded chromosome analysis, or from paraffin-embedded tissue and other specimens for which metaphase chromosomes could not be obtained.3–5 Limitations of CGH include the inability to detect balanced genomic rearrangements, and a limit of resolution of 3 to 5 Mb at best.6 Because of its vastly superior resolution, array-based CGH has essentially replaced metaphase-based CGH.
Multicolor FISH
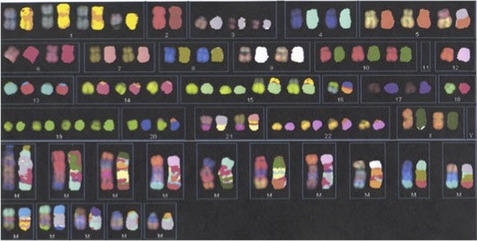
In the mid 1990s, techniques using multicolor whole chromosome paint probes that allowed simultaneous visualization of the 24 different human chromosomes in one hybridization experiment were introduced. The two main techniques, known as spectral karyotyping and multicolor FISH, involve combinatorial labeling of flow sorted chromosomes using different combinations of five fluorochromes.7,8 Spectral karyotyping utilizes spectral imaging and spectroscopy for image acquisition, whereas multicolor FISH utilizes a series of filters with defined emission spectra. These techniques have been helpful for the identification of marker chromosomes, although marker chromosomes are now often identified, and their content more precisely characterized, using array-based copy number assessment. Spectral karyotyping/multicolor FISH may still be useful for characterizing balanced interchromosomal rearrangements, particularly in neoplastic specimens that have poor banded morphology and complex rearrangements (Fig. 2); however, the limit of resolution is approximately 1 to 2 Mb for the detection of reciprocal terminal translocations.9 The use of these techniques in clinical laboratories is also limited by their expense and the time involved in performing the experiments and analysis.
Fiber FISH
The linear nature of metaphase chromosomes allows mapping of the position and orientation of sequences with respect to each other by FISH, but resolution is limited to probes that are at least 1 to 3 Mb apart.10 Interphase FISH provides much better resolution than metaphase FISH, but the DNA in this less condensed state is arranged in a random, nonlinear fashion. FISH on extended chromatin or DNA combines the qualities of both metaphase and interphase FISH in that it allows for high-resolution mapping on linear stretches of chromatin or DNA. This technique has been referred to by a variety of names, including FISH on free chromatin or DNA, FISH on extended or elongated chromatin or DNA fibers, FISH on DNA halos, and FISH on single or individual stretched DNA molecules, but the term “fiber FISH” has been used to encompass all of these variations.11 Fiber FISH mapping can distinguish probes separated by as little as 1 Kb on a DNA fiber.
Different methods have been developed for releasing extended chromatin or DNA from the nucleus, but subsequent FISH is similar to that performed on more conventional preparations.10 Methods for release include alkaline lysis or detergents, which is sometimes followed by mechanical stretching of the released fibers. DNA fibers can also be released from previously fixed cells, and molecular combing is a method that can be used to generate extended DNA from isolated DNA instead of lysed cells. Fiber FISH is not well-suited for use in the clinical laboratory owing to a number of factors, such as the time involved in preparing slides and nonstandard slide preparation; however, it has been used for a number of research applications that require high-resolution mapping. Some of these applications include determination of the size and structure of viral DNA integrated into a host genome; determination of the size, number, and orientation of amplicons within amplified regions of DNA; determination of the relationship and orientation of repetitive DNA elements (satellite sequences, arrays of alpha-satellites, etc); recognition of length differences in telomeres; and the study of the spatial and temporal distribution of DNA replication sites.
Flow FISH for the Determination of Telomere Length
Alteration of telomere length has been studied in the context of aging, cancer, and other disorders. A number of different techniques have been developed to measure telomere length.12 Analysis of terminal restriction fragment length by Southern blotting is reproducible but time consuming, requires a relatively large amount of DNA, and can overestimate the average telomere length by many Kb because of the distance between terminal restriction sites and the telomeric repeats. Polymerase chain reaction-based methods have also been used to determine telomere length, and these have their advantages and disadvantages as well. A quantitative FISH technique for measurement of telomere length on individual metaphase chromosomes has been developed that uses peptide nucleic acid probes that are complementary to single-stranded TTAGGG target sequences. Quantitative hybridization to telomere repeats is achieved by using conditions that allow peptide nucleic acid probes to hybridize to only the telomere repeat target sequences. This technique originally used image analysis, but was subsequently adapted for analysis of cells in suspension by flow cytometry (flow FISH). Additional modifications to the technique have included partial automation, inclusion of control cells with known telomere length, and the capability of limited immunophenotyping to analyze telomere length in subpopulations of leukocytes.
Although flow FISH is time consuming and technically demanding, it has been used in many research studies.12 Flow FISH is also being used as a diagnostic tool in patients who are suspected to have dyskeratosis congenita (DC), an inherited bone marrow failure syndrome that is characterized by short telomeres. Germline mutations in genes in the telomerase maintenance pathway are known to cause DC and other related disorders with short telomeres; however, mutations have only been identified in approximately half of the patients to date who meet clinical criteria for DC.13 Very short telomeres, defined as telomere length below the first percentile compared with healthy controls, have been found by flow FISH in leukocytes from individuals with DC.13,14 The finding of very short telomeres seems to have a high diagnostic sensitivity for DC, but the specificity for distinguishing DC from other bone marrow failure disorders differed between two studies.13,14 It is not known at present if this difference in specificity is real or owing to technical differences.
Oligonucleotide-Based High-Resolution FISH
FISH with oligonucleotide probes was originally limited to repetitive DNA targets, but recent advances in the de novo synthesis of long oligonucleotides (>150 mers) allow visualization of unique sequence targets at a resolution that has not been possible with probes generated from cloned DNA. Careful in silico selection of oligonucleotides for non-repetitive, unique genomic sequence generates high signal to noise without the need for suppression of repetitive DNA sequence. With selected, tiled oligonucleotide probes, Yamada and colleagues1 were able to visualize regions as small as 6.7 Kb by FISH using conventional cytogenetic preparations and standard FISH protocols. These investigators were also able to determine the genomic structure of the same 479-Kb duplication in 15q13.3 in two different individuals using a 2-color oligonucleotide probe set. The duplication was visualized as a direct duplication in one individual, whereas in the second individual the signal pattern was consistent with an inverted duplication. Thus, oligonucleotide FISH may provide an easier alternative to fiber FISH for high-resolution mapping of genomic structure in some situations. Oligonucleotide FISH would also be useful in the clinical laboratory for visualizing deletions and duplications detected by microarray that are below the level of resolution of FISH using bacterial artificial chromosome probes.
Applications of Interphase FISH
Technical Considerations
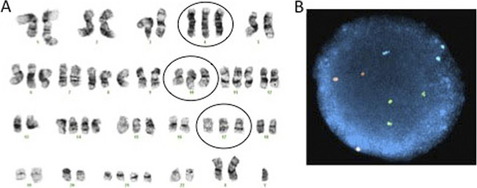
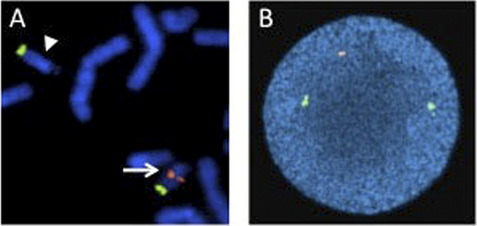
A significant advantage of interphase FISH over metaphase FISH is that actively dividing cells are not necessary; therefore, interphase FISH can be performed on a wider range of specimens. A much larger number of nuclei can also be scored very quickly by interphase FISH, allowing for the detection of low level mosaicism and minimal residual disease. For interphase FISH analysis, probe design or strategy for the identification of chromosomal abnormalities depends on the specific alteration to be detected. Enumeration probes consist of either centromeric/pericentromeric probes for detecting aneusomies, or unique sequence probes for detecting aneusomies, deletions, duplications, or amplification (Figs. 3 and 4). Fusion probe strategies are used for identifying specific translocations or inversions, and consist of probes from the two targets involved in the rearrangement that are labeled in two different colors (eg, red probe for PML at 15q24.1 and green for RARA at 17q21.2). In the normal state, the two probes are separated; however, the presence of the relevant rearrangement results in juxtaposition of the probes to give a fusion signal. First-generation fusion probes gave single fusion signals that can result in false-positive results in some proportion of interphase nuclei owing to coincidental overlap of probes. Dual-fusion probe designs circumvent this problem by creating two fusion signals, thereby essentially eliminating the possibility of false-positive signals owing to chance probe overlap (Fig. 5). Translocations and inversions can also be identified using a break-apart probe strategy, in which different color probes that flank the breakpoints of the rearrangement are utilized. In the normal state, the break-apart probe gives two fusion signals, whereas a rearrangement results in separation of the fused signal to give a red signal and green signal (Fig. 6). For informative schematic diagrams illustrating the dual fusion and break-apart probe strategies, refer to the review by Ventura and colleagues.15
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree