The Evolving Picture of Microdeletion/Microduplication Syndromes in the Age of Microarray Analysis: Variable Expressivity and Genomic Complexity
Department of Pathology, Duke University, Durham, NC USA
Keywords
• Microdeletion • Microduplication • Syndrome • Microarray • 1q21 • 15q13 • 15q24 • 16p13.1 • 16p11.2 • 17q21.3
Submicroscopic genomic copy number changes not visible by routine chromosome analysis are a common cause of congenital anomalies, developmental delay, and neuropsychiatric disorders. In previous decades, most recurrent genomic rearrangements were identified in a phenotype-first manner. In other words, a distinct collection of features and congenital anomalies distinguished a particular patient population, which was then interrogated for a common genetic anomaly. The clinical phenotypes of Prader-Willi and DiGeorge syndromes were described in 1956 and 1968, respectively, but the underlying genomic lesions were not described until 1981.1,2 The advent of genomic microarray testing has ushered in a new age of discovery in which the genomic change is often identified before a clear common clinical picture has emerged.
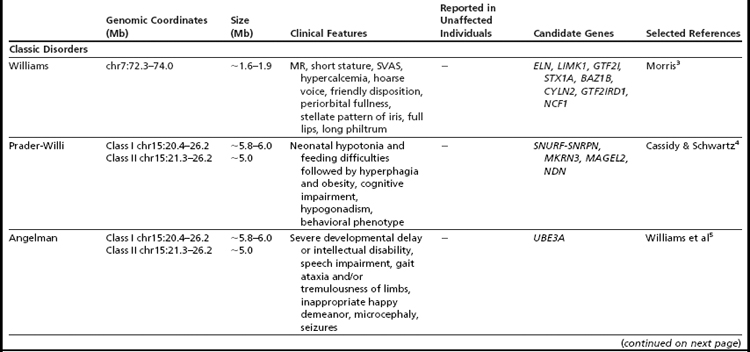
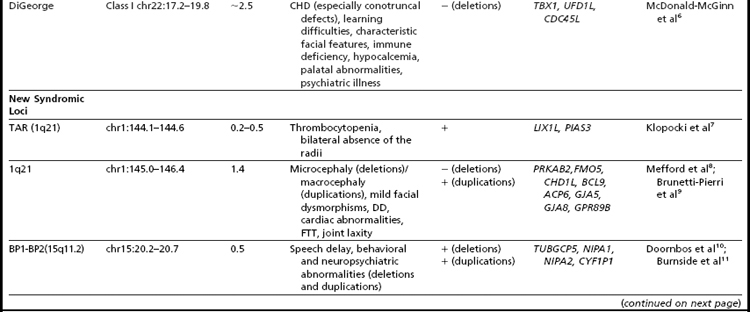
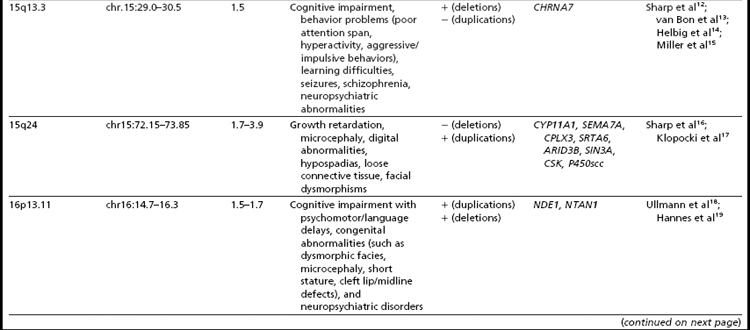
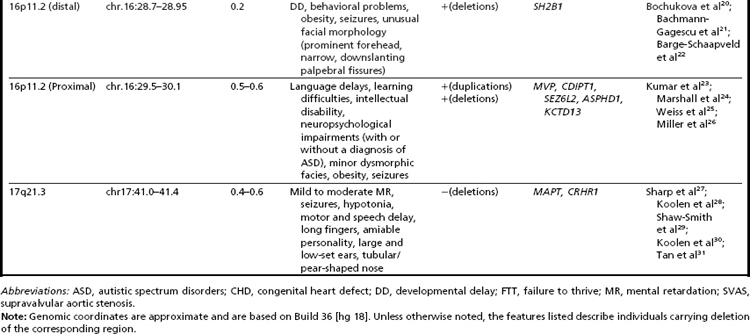
Many of the classic microdeletion syndromes, such as DiGeorge, Prader-Willi, Angelman, and Williams syndromes, for example, as well as many more-recently described deletions and duplications, are recurrent because of the underlying architecture of the genome (Table 1). Approximately 5% of the human genome comprises blocks of repetitive sequences greater than 1 kilobase (kb) in size that share 90% sequence homology with other regions of the genome.27 When these segmental duplications flank an intervening block of unique sequence, nonallelic homologous recombination (NAHR) can cause deletion, duplication, or inversion of that unique sequence. The internal structure and orientation of these repetitive blocks is polymorphic, and often a particular orientation such as an inversion facilitates NAHR. In this review, the authors focus on 9 regions of the genome that are flanked by repetitive sequences. Whereas the phenotypic consequences of aberrations in some of these regions are clear, the clinical significance of copy number changes in other regions remains to be fully elucidated. Many of the rearrangements discussed in this review and other recent reviews32,33 are members of the “susceptibility” genre, characterized by variable expressivity and reduced penetrance. By themselves these genomic changes may not be sufficient to cause a clinically abnormal phenotype; however, when combined with other genomic and environmental insults, the cumulative or epistatic effects may result in an abnormal phenotype.34
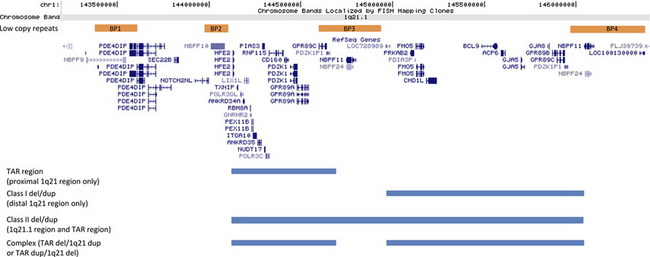
1q21.1
The genomic architecture of the 1q21 region predisposes it to NAHR-mediated rearrangements due to the presence of 4 segmental duplications of very high sequence identity, termed BP1-BP4, or break point 1, 2, 3…8 (Fig. 1). These regions of segmental duplication can serve as substrates for atypical recombination. Three deletions or duplications involving this region have been reported. The first aberration is deletion/duplication of the BP2-BP3 region that is associated with thrombocytopenia absent radius (TAR) syndrome.7 The distal 1.35 Mb deletion of 1q21.1, involving BP3 to BP4, is typically considered to be the classic 1q21.1 microdeletion syndrome.8 Deletions of BP2 to BP4 are often termed class II deletions and involve both the proximal TAR region as well as the 1.35 Mb distal 1q21.1 region and are approximately 2 Mb in total size.35
TAR Deletion/Duplication
TAR syndrome is characterized by the bilateral absence of the radius bone and a dramatically reduced platelet count or hypomegakaryocytic thrombocytopenia. The syndrome is associated with a microdeletion within 1q21.1, with a minimum deletion interval of 200 kb between BP2 and BP3 that is necessary, but not sufficient, to cause the phenotype.7 The most frequently observed deletion is roughly 500 kb (chr.1: 144.1–144.6 Mb, hg18) and contains approximately 17 known genes. Parental analyses have shown that this deletion is de novo in only 25% of affected individuals. Therefore, it has been postulated that the phenotype develops only in the presence of additional, although still unknown, modifiers.7 It is unclear whether reciprocal duplications of this region may have phenotypic consequences36 or represent benign variants, because they are sometimes seen in unaffected parents (KLD and CWR unpublished observation). Within the minimum interval, 2 candidate genes seem to be the most promising contributors to the phenotype seen in these individuals. The PIAS3 gene product has interactions with STAT3,37 a transcription factor involved in hematopoietic growth factor signaling, and the LIX1L gene shares sequence homology to the Lix1 gene, which is expressed during chick hind-limb development.38
1q21.1 Deletion
The 1q21.1 microdeletion involves minimally the material between segmental duplication blocks BP3 and BP4, which spans an approximately 1.35 Mb region (chr.1: 145.0–146.35 Mb, hg18).8 Clinical features are variable, and unlike some copy number changes, do not seem to lead to a clinically distinguishable phenotype. Typically, individuals with aberrations of 1q21 are discovered by microarray analysis, representing a genotype-first model of diagnosis. The most common clinical findings in these individuals are microcephaly, mild intellectual disability, dysmorphic facial features, and eye abnormalities. Many other features may also be present but are typically seen in less than 25% of individuals.8 These findings include seizures, skeletal malformations, cardiac defects, and genitourinary anomalies. Deletions of 1q21.1 are also considered to be a risk factor for psychiatric disorders such as autism spectrum disorder (ASD), attention deficit hyperactivity disorder, sleep disturbances, and schizophrenia.39
There are a total of 8 OMIM genes found in the 1.35 Mb region of 1q21.1: PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8, and GPR89B. Although no genotype-phenotype correlations have been described, 2 of these genes, GJA5 and GJA8, have been suggested to contribute to some of the clinical features seen in patients with 1q21.1 deletions. Mutations and deletions of the GJA5 gene have been identified in individuals with atrial fibrillation and structural cardiac defects.40,41 In a study of individuals with congenital heart defects, .6% of these patients were found to have deletions involving GJA5.40 Mutations in the GJA8 gene have been detected in cases of several types of cataracts as well as cataract-microcornea syndrome.42–46 Deletions of GJA5 and GJA8 are therefore hypothesized to play a role in the cardiac and eye abnormalities observed in individuals with 1q21.1 deletions.8 The BCL9 (B cell CLL/lymphoma 9) gene, a transcription factor in the Wnt signaling pathway, has been proposed as a candidate gene for schizophrenia because this signaling pathway influences neuroplasticity, cell survival, and neurogenesis.47
Class II Deletion
Larger deletions that involve both the TAR and the 1.35 Mb critical region are known as class II deletions. The clinical features of these individuals seem to be similar to those of the distal 1q21.1 deletions and can also be variable. These features include developmental delay, cardiac defects, genitourinary anomalies, and dysmorphic features.9,35 Complex deletions and/or duplications can also occur where an individual has a deletion of the TAR region and duplication of the distal 1q21.1 region9 or vice versa.36
1q21.1 Duplication
Duplications of the distal 1q21.1 region (BP2–BP3) have also been described.8,9 Like many reciprocal duplications of microdeletion syndromes, the overall incidence seems to be lower, which is thought to be due to ascertainment bias because the duplication is associated with a less severe phenotype and therefore may not come to clinical attention. Common clinical features of 1q21.1 microduplications include dysmorphic features, developmental delay and intellectual disability, and autistic features, although the duplication has been observed in control cohorts.8 Unlike the 1q21.1 microdeletions where microcephaly is consistently seen, macrocephaly seems to be a prominent feature of the 1q21.1 microduplications. It is thought that one or more dosage-sensitive genes, related to the size and the development of the brain, are located in this region.9,48
Chromosome 15
15q13.3 Deletion/Duplication
The genomic architecture of chromosome 15 is complex (Fig. 2). Six regions of low copy repeats have been identified and are commonly referred to as break points 1 to 6 (BP1–6). Prader-Willi and Angelman syndromes are caused by deletions mediated proximally by either BP1 or BP2 and distally by BP3. More distally, deletions between BP4 and BP5 in the 15q13.3 region have also been reported. First described in a series of 5 probands identified by a whole-genome microarray screen of patients with mental retardation and/or congenital anomalies,12 additional studies13–15 have widened the phenotypic spectrum. In the cohort of probands described by van Bon and colleagues13 (N = 15), 14 displayed some degree of cognitive impairment ranging from mild learning problems to severe mental retardation. In that study, the only other features observed in over 50% of the probands were behavior problems such as poor attention span, hyperactivity, and aggressive and impulsive behavior. No common dysmorphic features were noted. When additional family members were examined to determine inheritance, 13 relatives with no cognitive deficits carrying the deletion were identified. Other carrier family members displayed abnormal phenotypes ranging from mild mental retardation to learning difficulties. Other studies have shown that deletion carriers demonstrate an increased frequency of seizures,14 schizophrenia,39 and other neuropsychiatric abnormalities.15 The deletion is thought to account for .24% of cases of idiopathic mental retardation,13 1% of cases of idiopathic generalized epilepsy,14 and .2% of cases of schizophrenia with only a general population frequency of less than .02%.39 However, given the wide interfamily and intrafamily variability in the phenotype, it seems unlikely that the deletion, in isolation, is sufficient to cause an abnormal phenotype. It has been hypothesized that some individuals possess the ability to outgrow the abnormal phenotype such that they display learning disabilities at a young age but grow to become normal functioning adults.13
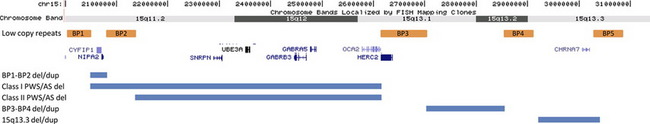
Fig. 2 The repetitive sequence blocks of proximal 15q and related disorders. Based on Build 36 [hg 18].
The BP4-BP5 region (chr.15: 29.0–30.5 Mb, hg18) contains 7 known genes: ARHGAP11B, MTMR15, MTMR10, TRPM1, KLF13, OTUD7A, and CHRNA7. CHRNA7 encodes the human alpha-7 neuronal nicotinic receptor subunit, which is a member of a family of ligand-gated ion channel proteins that mediate fast signal transmission at synapses. This gene is the most thoroughly investigated candidate gene in the region. Outside of microdeletion studies, polymorphisms in the gene have been linked to schizophrenia49 and epilepsy,50 and the knockout mouse displays an abnormal EEG,51 making CHRNA7 a promising contender as an important causative gene.
Duplication of the BP4-BP5 region has also been reported.13,15,52 These duplications were found in individuals being investigated because of developmental delay/mental retardation, multiple congenital anomalies, dysmorphic features, autism or autistic spectrum, or seizures (typical indications for chromosomal microarray analysis). The clinical significance of the BP4-BP5 microduplications as well as smaller duplications of the CHRNA7 gene is unclear, because many families have both affected and unaffected individuals who carry the duplication. It too may be a risk factor for neuropsychiatric disease.
BP1-BP2 Deletion/Duplication and Other Abnormalities in the Region
Because of the unique structure of chromosome 15, additional deletions and duplications have been reported that use atypical combinations of the break points. Van Bon and colleagues13 described one family in which a BP3-BP4 deletion failed to segregate with an abnormal phenotype supporting the characterization of this deletion as a benign variant. Subsequent studies have suggested that although not sufficient to cause an abnormal phenotype, BP3-BP4 deletions could be contributory. Common features cited include failure to thrive, hypotonia, renal anomalies, and premature puberty, although variable expressivity and inheritance of the deletion from a “normal” parent were also documented.53 Maternally derived deletions mediated by BP1 or BP2 proximally and BP4 or BP5 distally have been shown to result in a more severe Angelman phenotype, with these individuals scoring lower on assessments across all areas of mental, social, and language development.54 In fact, some studies have reported phenotypic differences between patients with Prader-Willi syndrome depending on the proximal deletion break point, BP1 (type 1) or BP2 (type 2).55
Recently, two studies have reported cohorts of patients (N = 910 and N = 14611) with deletions and duplications mediated proximately by BP1 and distally by BP2. Both studies indicate that phenotypic consequences of both deletion and duplication of this region are highly variable. The abnormal features most significantly associated with deletion were behavioral/neuropsychiatric anomalies and speech delay, observed in 67% and 92% of evaluated individuals, respectively.11 Some probands showed inheritance of the deletion or duplication from an apparently normal or mildly affected parent; however, most parents did not undergo rigorous behavioral or psychiatric evaluation. The authors of both studies acknowledge that their data could be skewed by ascertainment bias and/or small control populations, but both advocate that copy number changes in this region could contribute to an abnormal phenotype. Additional investigation including a control study in which participants undergo in-depth psychiatric testing is recommended. Four genes are contained in the region, TUBGCP5, NIPA1, NIPA2, CYF1P1. These genes are highly conserved, not imprinted, and may play roles in cognition and behavior.10,11,55,56
A third study that specifically examined candidate loci in a large cohort of patients with idiopathic generalized epilepsies reported BP1-BP2 deletions in 1% of their patient population (n = 1234) and in .2% of their control population.57 Although enriched in the affected population, the deletion failed to segregate with the seizure phenotype in 3 large families, emphasizing the fact that other factors (stochastic events, environmental effects, recessive mutations on the other allele, and background genomic variation) must play a role in the phenotypic expression of this deletion.
15q24 Deletion
Sharp and colleagues16 first described a cohort of 4 patients, all with mild to moderate developmental delay, growth retardation, hypospadias, microcephaly, digital abnormalities, and joint laxity, who were found to have microdeletions of the 15q24 region. The investigators suggested that individuals with these deletions could share enough common features to potentially represent a clinically recognizable syndrome. To date, few additional patients have been described to both clarify the clinical presentation and to delineate the smallest region of overlap (SRO) for the 15q24 microdeletion syndrome. The reported phenotypic features of individuals with deletions of 15q24 have consistently included developmental delay that is mild to moderate, growth retardation or short stature, hypotonia, digital abnormalities, skeletal deformities such as joint laxity, genital abnormalities, and characteristic facial features. 7,58–61
NAHR has been proposed as the mechanism underlying these rearrangements because of the presence of 3 highly identical segmental duplications located within this region, and the majority of the described 15q24 deletions do have break points in these repetitive regions. The discovery of additional repetitive elements in the region has come from the analysis of alternative break points in newly described individuals.58 The total size of the 15q24 deletions range from approximately 1.7 to 6.1 Mb, and the current SRO has been defined as an approximately 1.2 Mb region between break points termed 15q24B and 15q24C.17,61 This region is gene-rich, and genes such as CYP11A1, SEMA7A, CPLX3, SRTA6, ARID3B, SIN3A, and CSK have been proposed to contribute to the phenotype.
15q24 Duplication
One patient with duplication of the 15q24 region has been described by Kiholm Lund and colleagues.62 Although the duplication was inherited from a healthy father, features similar to that of the 15q24 microdeletion syndrome were noted, such as global developmental delay and dysmorphic features, along with digital and genital abnormalities. One additional case with 15q24 microduplication that encompasses the SRO for the reciprocal microdeletion has been documented. A male with mild mental retardation, decreased joint mobility, digital abnormalities, and characteristic facial features was described.58 He was also described as having attention deficit hyperactivity disorder and Asperger syndrome. Cukier and colleagues63 identified a small 10-kb duplication located within the 15q24 critical region, found to be inherited identical by descent in first cousins with autism. Because this duplication encompasses only a single gene, UBL7, it was suggested that these individuals effectively narrowed the critical region for susceptibility to the development of ASDs in 15q24 rearrangements.
Duplications adjacent and distal to the minimal critical region for the 15q24 deletions have also been described in 2 families. These duplications are also likely to be NAHR-mediated because of the abundance of low copy repeats located in this region. In both studies, 3 individuals are described including 2 siblings and their mother. All individuals exhibited mild developmental delay, hypotonia, digital abnormalities, and characteristic facial features.58,64 Because of the lack of described cases, it remains unclear if 15q24 duplications, especially those outside of the SRO described in the 15q24 deletions, result in a distinct clinical phenotype.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree