Healthcare-Associated Respiratory Viral Infections
Wing Hong Seto
Janice Lo
INTRODUCTION
Acute respiratory viral infections are frequent causes of hospital admission, and healthcare-associated infection clusters of these diseases are frequent occurrences. Despite the large number of viruses causing these infections and the large number of admissions with such infections, the size of the problem worldwide is not known. Determining the extent of this problem is complicated by the lack of laboratory diagnostic capabilities. However, in recent years, many hospitals have established rapid viral diagnosis capacity, which should be set up if it is still unavailable. In Hong Kong, such capacity has existed since 1995. The number of viral laboratory diagnoses made for the year 2011 in the Hong Kong government virology laboratory is shown in Table 42.1. As shown, influenza and respiratory syncytial virus (RSV) account for the majority of positive specimens and together account for about 70%. The majority of these patients are children and in adults; many are elderly. It is obvious that the practice of infection control for these patients has its own particular challenges and demands.
This chapter is divided into two sections. The first section provides vital background information on the viruses as it pertains to infection control and the second covers the strategies for infection control.
THE VIRUSES
Although deaths from acute viral respiratory illness gradually decreased until they are now uncommon in developed countries, the morbidity associated with respiratory illness continues to be a major societal burden. The large number of infectious agents involved makes successful vaccination difficult and development of antivirals for the treatment of these infections slow. Implementation of nonpharmaceutical intervention is essential to limit the spread of acute viral respiratory pathogens in hospital settings and the community.
TABLE 42.1 Viruses (Patient-Specific) Identified in Hong Kong Government Virology Laboratory in 2011 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Most of the respiratory viral pathogens are subject to marked seasonal influence. Seasonality for a specific agent may be different for different regions, especially when the climates are different. Although the underlying mechanisms still remain largely unexplained, the seasonality for many infectious diseases is stable and well-documented (1,2). The knowledge of seasonal and cyclical variations in the incidence of diseases is not only useful in clinical management of patients but also essential for the design and implementation of preventive strategies.
To devise effective infection control measures, it is essential to understand the factors that affect the spread of respiratory virus pathogens, which include:
Virus concentrations in respiratory secretions
Duration of virus shedding
Ability of the virus to survive in aerosols or on the surface of hands or the inanimate environment
Route of infection
Minimal infectious dose
Innate and specific host immunity
Social factors (e.g. crowding, mobility of people)
Viruses causing respiratory disease comprise a number of agents from different families, including Orthomyxoviridae (human influenza viruses), Paramyxoviridae (human parainfluenza viruses, RSV, and human metapneumovirus), Coronaviridae (human coronaviruses and severe acute respiratory syndrome-related coronavirus), Adenoviridae (human adenoviruses), and Picornaviridae (human rhinoviruses). These viruses may infect different parts of the respiratory tract, giving rise to various symptoms, from rhinorrhea and sore throat in the upper respiratory tract to bronchiolitis and pneumonitis in the lower respiratory tract. The degree of severity of the clinical features may range from subclinical to severe morbidity and mortality. The same virus may have different clinical presentations in different individuals, and host factors and immunologic status play important roles in modifying the illness.
Clinical diagnosis cannot reliably differentiate between infections with these various pathogens. Definitive diagnosis requires laboratory investigations. The recent increasing use of molecular testing for a panel of respiratory viruses has reduced the turnaround time for laboratory results as compared with conventional viral culture or antigen detection tests, while
providing high analytical sensitivity. Nevertheless, a definitive diagnosis of the etiologic agent is not mandatory for patient management, the mainstay of which is supportive treatment with symptomatic relief, and infection control measures mainly in the form of droplet precautions. This is in line with the route of transmission of respiratory viruses by droplet spread and contact of respiratory mucous membranes with infectious secretions. Regarding disinfection of contaminated surfaces, the agents effective against both enveloped and nonenveloped viruses should be used, such as sodium hypochlorite freshly prepared at the appropriate concentration. Only occasionally would a specific laboratory diagnosis be clinically relevant, such as support for the continued use of antiviral therapy in patients with influenza, and confirmation of the diagnosis in severe cases or immunocompromised patients. Laboratory confirmation of the etiology of outbreaks of respiratory illnesses also is useful to guide control measures. On a public health scale, laboratory data is helpful in defining the epidemiology of various viral infections in a locality, in terms of affected age groups, seasonality, and cyclical patterns.
providing high analytical sensitivity. Nevertheless, a definitive diagnosis of the etiologic agent is not mandatory for patient management, the mainstay of which is supportive treatment with symptomatic relief, and infection control measures mainly in the form of droplet precautions. This is in line with the route of transmission of respiratory viruses by droplet spread and contact of respiratory mucous membranes with infectious secretions. Regarding disinfection of contaminated surfaces, the agents effective against both enveloped and nonenveloped viruses should be used, such as sodium hypochlorite freshly prepared at the appropriate concentration. Only occasionally would a specific laboratory diagnosis be clinically relevant, such as support for the continued use of antiviral therapy in patients with influenza, and confirmation of the diagnosis in severe cases or immunocompromised patients. Laboratory confirmation of the etiology of outbreaks of respiratory illnesses also is useful to guide control measures. On a public health scale, laboratory data is helpful in defining the epidemiology of various viral infections in a locality, in terms of affected age groups, seasonality, and cyclical patterns.
INFLUENZA VIRUSES
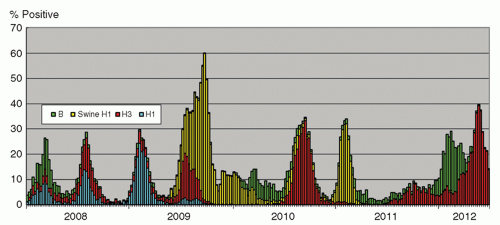
The influenza virus, an enveloped RNA virus, is classified into types A, B, and C. It is considered as highly significant among agents causing respiratory viral infections. The infection ranges from upper respiratory tract involvement with a typical “influenza-like illness” with fever, myalgia, cough, and sore throat, to lower respiratory tract disease with pneumonitis, and occasionally severe systemic involvement. The incubation period usually is 1 to 3 days, and the virus is shed in respiratory droplets and secretions from clinical onset for a few days, and longer for children until up to a week (3). Virologically, the genome of influenza A and B viruses comprises eight RNA segments. Two of the genes, hemagglutinin (H) and neuraminidase (N), encode for surface proteins targeted by the immune system. Influenza A viruses comprise 17 designated H subtypes and 9 N subtypes. Viruses of subtypes H1 to 16, combined with different permutations of N subtypes, can be found in avian species as natural reservoirs of the viruses (4). The H17 virus was initially reported in 2012 from bats in Guatemala (5). Other animals, including humans, harbor a more limited number of subtypes of influenza A viruses. As a result of the replacement of entire gene segments, especially of the H and N genes of the influenza A virus, due to recombination events in nature, called genetic shift, viruses for which human has little immunity emerge periodically, causing pandemics (6). In the 20th century, successive influenza A pandemics have been caused by H1N1 (1918), H2N2 (1957), H3N2 (1968), and most recently H1N1 (2009). Pandemics have different characteristics in terms of morbidity and mortality rates, and the predominantly affected age groups and populations. This probably is due to a combination of viral and host immunologic factors, and the availability of medical and supportive treatment, including the use of antimicrobials for secondary bacterial infections, the development of antiviral agents, advances in life-support measures, and heightened infection control practices. The 1918 pandemic was caused by a virus likely to be entirely of avian origin (7), with around 30% of the world’s population being infected, and a mortality of 40 million people, especially among young adults (8). Secondary bacterial infections probably played a significant role (9). The 1957 and 1968 pandemics had lower mortality, probably as a result of a lesser degree of virulence of the virus, and improved medical management (10). The virus causing the 2009 pandemic likely had a swine origin (11). The overall mortality was <0.5%. In contrast to seasonal influenza, most of the serious illnesses caused by the 2009 pandemic virus have occurred among children and non-elderly adults, and ˜90% of the deaths have occurred in those <65 years of age (12). This phenomenon is postulated to be due to the related H1N1 virus having circulated from 1918 until 1950s, so that those born during this period had a degree of protective immunity due to previous infection (13). Apart from antigenic shift, influenza A and B viruses undergo antigenic drifts where cumulative mutations mainly in the H gene give rise to emergence of new strains against which there is reduced immunity in the population (14). Such strains cause outbreaks of various severity and durations in different seasons. The seasonality of influenza infections is distinct in different areas of the world; countries in temperate climates mainly have winter peaks, and tropical areas have a more even distribution throughout the year. In Hong Kong, there are two distinct peaks in winter and in summer in most years, although variations may be encountered (Figure 42.1). Influenza C causes mild sporadic infections and is not of major medical concern.
Surveillance of influenza A and B infections, both clinical and laboratory-based, is of significance on a global scale, in order to monitor for the occurrence of antigenic drift and
shifts. The World Health Organization (WHO) has a laboratory network where virus strain characterization is systematically undertaken by national and collaborating reference laboratories, in order to monitor the currently circulating influenza strains, and to make recommendations on a half-yearly basis on the strains to be included in the vaccine for the Northern and Southern hemispheres (15).
shifts. The World Health Organization (WHO) has a laboratory network where virus strain characterization is systematically undertaken by national and collaborating reference laboratories, in order to monitor the currently circulating influenza strains, and to make recommendations on a half-yearly basis on the strains to be included in the vaccine for the Northern and Southern hemispheres (15).
Novel Influenza Viruses (Including Avian Influenza)
Apart from the circulating seasonal influenza strains, sporadic infections of influenza A strains crossing the species barrier to infect humans are reported regularly, mainly from avian and swine origin. Since 1997, human infections with the highly pathogenic influenza A H5N1 virus from avian origin have been detected repeatedly in different countries. The overall mortality rate of the infection is >40% (16). Various genetic characteristics of the virus with putative association with virulence have been reported (17). The incubation period of influenza A H5 infection usually is 2 to 5 days, and the virus has a predilection for infecting the lower respiratory tract, apparently related to the presence of receptors primarily binding the virus (i.e., sialic acid bound to galactose by α-2,3 linkages) (18). In the infected patient, virus shedding occurs from the respiratory and gastrointestinal tract possibly for >1 week, although the course is still incompletely characterized (19). So far, clusters of human infection with likely human-to-human transmission have been limited to exposure to infected respiratory secretions in blood relatives, implying host predisposing factors to infection (19,20). Nevertheless, vigilance is important lest the virus maintains its virulence on crossing the species barrier to result in efficient transmission among humans (21,22). Influenza A H9N2 infections also have been occasionally detected in humans as a result of direct transmission from avian origin. The infection is mostly mild and self-limiting (23). Virus strain surveillance on the global scale covers avian viruses as well, and has yielded candidate seed viruses for vaccine development in anticipation of emergence of new viruses with pandemic potential (24).
Swine influenza A viruses also have occasionally been detected in human infections. The disease has been mostly self-limited and similar to seasonal influenza viruses. Nevertheless, the pandemic potential exists. In fact, the 2009 pandemic H1N1 virus has also been demonstrated genetically to be most closely related to swine viruses (11). In 2011, mostly sporadic episodes of human infections with swine-origin influenza A viruses were detected in the United States. Intensified surveillance of the strain so far has not yielded evidence of sustained human transmission (25).
Antivirals for Influenza
Another important aspect of influenza virus surveillance is the monitoring of the emergence and prevalence of antiviral resistance. Currently, there are two major groups of antivirals active against influenza: the adamantanes, which may be effective against influenza A but not influenza B, and the neuraminidase inhibitors, which may be effective against both influenza A and B. The 2009 pandemic influenza A H1N1 virus and the currently circulating H3N2 virus are resistant to adamantanes and generally susceptible to neuraminidase inhibitors. Nevertheless, occasionally, mutation in the N1 gene results in an amino acid change (H275Y) and confers resistance of the H1N1 virus to the most widely used neuraminidase inhibitor, oseltamivir, whereas circulating influenza A H3N2 and influenza B viruses remain largely susceptible (15).
HUMAN PARAINFLUENZA VIRUSES
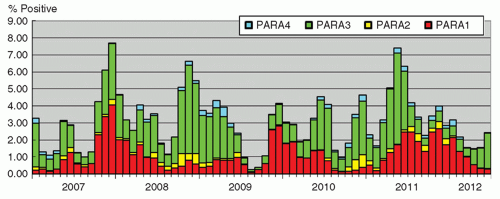
Human parainfluenza viruses (HPIVs) are enveloped RNA viruses, and are classified into types 1, 2, 3, and 4. They are the etiologic agents of various respiratory tract infections, especially in children, such as croup (laryngotracheobronchitis), which is more commonly associated with HPIV-1 and -2, and bronchiolitis, occurring mainly from infection with HPIV-3. The disease is usually self-limited, with occasional episodes of severe disease probably as a result of host factors and compromised immunity. The infection is mainly recognized in children, although the elderly are occasionally affected (26). The incubation period ranges from 2 to 6 days, and virus shedding occurs via infective respiratory secretions for around 1 week (14). Seasonal patterns are evident, with HPIV-1 and -2 occurring mainly in autumn, with HPIV-1 infections showing a cyclical peak every alternate year, whereas HPIV-3 mainly occurs during the summer months (27). The seasonal pattern in Hong Kong is shown in Figure 42.2. A definitive diagnosis for the infection may not be essential since a syndromic approach to patient management and infection control usually suffices.
RESPIRATORY SYNCYTIAL VIRUS
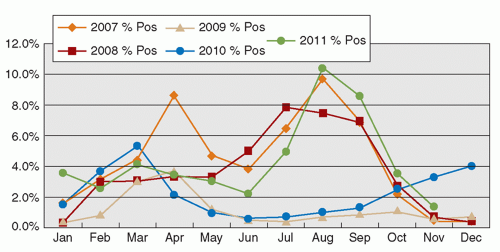
RSV is an enveloped RNA virus, classically causing bronchiolitis in young children <2 years of age. The infection may predispose to future asthmatic attacks. The infection typically peaks during the winter season in temperate climates. Nevertheless, the seasonal pattern varies geographically. In Hong Kong, the virus activity occurs mainly in the spring and summer months of March to September (28). Interestingly, with the high activity of the influenza A H1N1 in 2009 and 2010, the RSV peaks were abrogated (Figure 42.3). The incubation period of the infection is 4 to 5 days, and the virus is shed from infective respiratory secretions several days to >1 week (14). Since the disease mainly occurs in infants and young children with symptoms of lower respiratory tract infection, hospital admission is common. Outbreaks of RSV infection in pediatric hospital wards, and also in institutional settings, including those for the elderly, are commonly encountered. With a definitive laboratory diagnosis, treatment with ribavirin may be administered, especially in severe and immunocompromised patients. Scrupulous implementation of droplet precautions in all episodes of respiratory tract infections in the hospital setting, with environmental decontamination using disinfectants active against these viruses, is effective in preventing outbreaks and transmission of the pathogen.
HUMAN METAPNEUMOVIRUS
The human metapneumovirus (HMPV), an enveloped RNA virus, was first described in 2001 (29), and now is recognized as one of the viral agents causing respiratory tract infections. The incubation period is 5 to 6 days (30), and shedding of the virus can be for 1 to 2 weeks (31). The symptoms range from mild upper respiratory tract disease to lower respiratory tract and pulmonary involvement. The infection is observed in all age groups, with a high prevalence in pediatric patients. Seasonality studies suggest peak of infection occurring 1 to 2 months later than that for RSV epidemics (31). Occasionally, HMPV is shown to be the cause of clusters of respiratory tract infection in the community, but does not appear to be an important agent of hospital outbreaks.
CORONAVIRUSES
Human coronaviruses have been described since the 1960s to be the agents of the common cold. The viruses initially recognized were OC43 and 229E (32). Sporadic and self-limited infections usually were encountered, and there was not a great deal of medical interest, compounded by the lack of readily accessible diagnostic tests. It was in 2003 that the coronavirus came under global limelight with the occurrence of severe acute respiratory syndrome (SARS).
SEVERE ACUTE RESPIRATORY SYNDROME
The disease syndrome was first reported in November 2002 in Guangdong, China (33). The occurrence of infections at the level of community outbreaks first came to global attention in February 2003 in the city of Guangzhou. Epidemiologic investigations subsequently traced the spread of the virus, via Hong Kong, with extensive outbreaks to various countries including Vietnam, Singapore, and Canada. The outbreak of SARS was officially over in July 2003 as announced by the WHO. There were >8,000 infected patients, resulting in almost 800 deaths in 27 countries in a global scale (34). In Hong Kong, the infection count was 1,755 patients and 299 deaths. Nearly a quarter of the infected patients were healthcare workers. At the height of the outbreak, the virus was found to be a coronavirus first recognized to occur in humans, and the virus was named SARS-related coronavirus (SARS-CoV) (35). With genetic and epidemiologic studies, the virus was traced first to masked palm civets in game animals markets in Guangzhou, and eventually to bats as the probable natural reservoirs (36). The SARS-CoV is distinct in that the infective course was typified by shedding of low concentrations of the virus in the first week of infection, with peak shedding occurring in the second week (37). This shedding pattern enables the opportunity for infection control measures to be successfully implemented on early recognition of infection, probably contributing to the eventual successful control of the disease. In addition, there is evidence that the hygienic measures implemented on a population level during SARS resulted in a reduction in the incidence of other respiratory viruses (38). Although the SARS-CoV has now been eliminated from
circulation in humans, its recurrence is potentially possible, not only as a result of reintroduction from its natural reservoir but also of inadvertent escape from the laboratory (39).
circulation in humans, its recurrence is potentially possible, not only as a result of reintroduction from its natural reservoir but also of inadvertent escape from the laboratory (39).
NEWER CORONAVIRUSES
Since 2003, there was heightened interest in coronaviruses, and an explosion in publications on the agent. New coronaviruses infecting humans were subsequently recognized, including NL63 and HKU1 (40,41,42). These agents usually cause sporadic mild respiratory tract infections, similar to the earlier coronaviruses, and were much less virulent than the SARS-CoV.
HUMAN ADENOVIRUSES
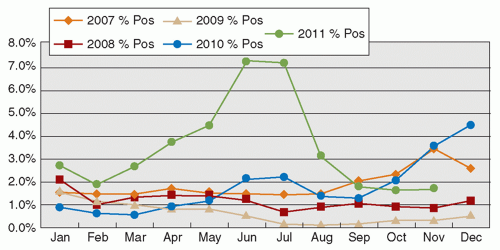
Human adenoviruses (HAdVs) are nonenveloped DNA viruses, now with >50 types recognized. HAdVs cause a plethora of diseases not limited to the respiratory tract. Respiratory adenoviruses mainly belong to types 1 to 7, and occasionally to other types. Mild upper respiratory illness is the usual presentation, although occasional episodes of pneumonitis are recognized (14). Although infection with the virus is mostly recognized in children, both young adults and the elderly can be infected. Outbreaks of HAdV infection are commonly reported in the community, with HAdV types 4 and 7, and more recently type 14, being associated with clusters of infection among military recruits (43). In recent years, new types of HAdVs continue to be described, and occasional outbreaks have been recognized (44). The seasonal occurrence of HAdV infection is not consistently reported in the literature. The seasonal pattern observed in Hong Kong is illustrated in Figure 42.4.
HUMAN RHINOVIRUSES
Human rhinoviruses are nonenveloped RNA viruses comprising >100 serotypes. They are classical causes of the common cold and are associated with mild respiratory disease. Episodes of severe infection seldom occur and are mostly confined to immunocompromised hosts (45). In terms of seasonality, infections more commonly occur from autumn to spring in temperate climates, whereas in the tropics, the incidence is highest during the rainy season (3). Distinct outbreaks are not commonly recognized. Nevertheless, due to the large number of serotypes, reinfections throughout life are frequent. The virus is mostly community-based, and rarely a problem in the hospital setting.
STRATEGIES FOR INFECTION CONTROL
UNDERSTANDING THE MODES OF TRANSMISSION
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree