Technological advances in protein and genetic analysis have altered the means by which platelet disorders can be characterized and studied in health and disease. When integrated into a single analytical framework, these collective technologies are referred to as systems biology, a unified approach that links platelet function with genomic/proteomic studies to provide insight into the role of platelets in broad human disorders such as cardiovascular and cerebrovascular disease. This article reviews the historical progression of these applied technologies to analyze platelet function, and demonstrates how these approaches can be systematically developed to provide new insights into platelet biomarker discovery.
Key points
- •
Technological advances in protein and genetic analysis have altered the means by which platelet disorders can be characterized and studied in health and disease.
- •
When integrated into a single analytical framework, these collective technologies are referred to as systems biology, a unified approach that links platelet function with genomic/proteomic studies to provide insight into the role of platelets in broad human disorders such as cardiovascular and cerebrovascular disease.
- •
Analyzing and comparing genetic expression data with sophisticated proteomic identification technologies has altered the landscape for delineating gene/protein networks regulating functional platelet responses.
Introduction
Relevance of Platelet Functional Disorders in Health and Disease
Molecular platelet disorders cause bleeding syndromes of varying severity, best characterized for cell-surface receptor defects involving α IIb β 3 and the GPIb-IX-V complex, and less frequently known to involve distinct signaling pathways or defects of granule formation. By contrast, there is a paucity of information on the molecular etiology of the “prothrombotic” platelet, despite long-standing evidence for the role of activated platelets in the development of focal ischemia. The latter observation is clearly supported by clinical studies of platelet activation markers during transitory and thrombotic vascular events. Causal evidence for the critical influence of platelets in human vascular events is highlighted by the efficacy of antiplatelet agents as therapeutic and prophylactic modalities in the setting of cerebrovascular ischemia, as established by observations of fewer nonfatal strokes in patients with prior stroke or transient ischemic attack, and fewer nonfatal strokes among patients treated for completed stroke. Comparable cardiovascular data exist demonstrating that interventions with antiplatelet agents offer therapeutic efficacy as interventional strategies for coronary artery disease (CAD), acute coronary syndromes, and percutaneous transluminal coronary angioplasty, either in acute settings or as prophylactic modalities. Antiplatelet agents, however, are not without toxicity, and “breakthrough” thromboembolic episodes remain prevalent. Thus the development of risk stratification strategies and/or platelet phenotypic/genotypic assays (“biomarkers”) able to predict responsiveness or adverse events have emerged as a research direction of the biopharmaceutical industry, and have the potential for widespread clinical application.
Introduction
Relevance of Platelet Functional Disorders in Health and Disease
Molecular platelet disorders cause bleeding syndromes of varying severity, best characterized for cell-surface receptor defects involving α IIb β 3 and the GPIb-IX-V complex, and less frequently known to involve distinct signaling pathways or defects of granule formation. By contrast, there is a paucity of information on the molecular etiology of the “prothrombotic” platelet, despite long-standing evidence for the role of activated platelets in the development of focal ischemia. The latter observation is clearly supported by clinical studies of platelet activation markers during transitory and thrombotic vascular events. Causal evidence for the critical influence of platelets in human vascular events is highlighted by the efficacy of antiplatelet agents as therapeutic and prophylactic modalities in the setting of cerebrovascular ischemia, as established by observations of fewer nonfatal strokes in patients with prior stroke or transient ischemic attack, and fewer nonfatal strokes among patients treated for completed stroke. Comparable cardiovascular data exist demonstrating that interventions with antiplatelet agents offer therapeutic efficacy as interventional strategies for coronary artery disease (CAD), acute coronary syndromes, and percutaneous transluminal coronary angioplasty, either in acute settings or as prophylactic modalities. Antiplatelet agents, however, are not without toxicity, and “breakthrough” thromboembolic episodes remain prevalent. Thus the development of risk stratification strategies and/or platelet phenotypic/genotypic assays (“biomarkers”) able to predict responsiveness or adverse events have emerged as a research direction of the biopharmaceutical industry, and have the potential for widespread clinical application.
Clinical features
Genetic Risk Factors for Platelet-Associated Cerebrovascular/Cardiovascular Diseases
Research designed to identify genetic risk factors in patients with thrombophilia have shown the strongest association in venous thromboembolic disease, with less consistent results in subjects with cerebrovascular or cardiovascular disease. Evidence exists that abnormal expression of platelet proteins may favor platelet activation and thrombus formation, data best developed for platelet cell-surface glycoprotein receptors studied within the context of patients with CAD. For example, quantitative expression of the platelet integrin α 2 β 1 is variable, and platelets expressing higher levels have an increased ability to bind collagen. Similarly, the 807T/873A α 2 β 1 polymorphism is known to increase the surface density of receptors on platelets, and is associated with an increased risk of ischemic heart disease in homozygotes. This genetic risk is enhanced in the background of subjects who smoke, and may also represent a risk factor for stroke in young patients. Similarly, it has been suggested that the PL A1 GPIIIa (β3) polymorphism is associated with stroke in patients younger than 50 years, and that a Kozak T/C polymorphism within the GPIb gene is associated with stroke. Nonetheless, other studies have been unable to demonstrate a risk between common platelet single-nucleotide polymorphisms (SNPs) and thrombotic risk. Many of the initial attempts to dissect out genetic risk factors were largely case-control studies in which subjects were grouped based on known SNPs of well-characterized platelet protein receptors (eg, Pl A1 vs Pl A2 ); that is, candidate genetic studies. Indeed, an emerging paradigm is that platelet-associated vascular risk is regulated by a large number of genetic loci each exerting small effects; this concept is not dissimilar to highly heritable quantitative traits such as height.
Genetics of Functional Platelet Responses
Data using multiple activation models clearly demonstrate that platelet responses to the majority (if not all) agonists is highly variable within the population. By contrast, the extent of platelet responsiveness within individuals is consistent over time, unrelated to agonist or quantitative outcome measures. These observations are consistent with those that demonstrate high levels of platelet function heritability in siblings, twins, and families with a history of premature CAD. Platelets retain megakaryocyte-derived mRNA and an abundant and diverse array of microRNAs (miRNAs), and have evolved unique adaptive signals for maintenance of genetic and protein diversity. Quiescent platelets generally display minimal translational activity, although maximally activated platelets retain the capacity for protein synthesis. Newly formed “reticulated” platelets retain larger quantities of mRNAs and have been associated with enhanced thrombotic risk in patients with thrombocytosis. Approaches to study platelet functional responses as quantitative trait loci linked to overall platelet responsiveness is a logical extension of these collective observations, designed to integrate transcriptomic and genomic information to variability in platelet activation and disease outcome. Indeed, in the only study reported to date focusing on a 97-member candidate gene subset to analyze adenosine diphosphate (ADP) and collagen responses, the combined effects of multiple SNPs accounted for 38% to 46% (collagen) and 13% to 16% (ADP) of the platelet variability in a healthy cohort. This variability appears to be considerably less than that predicted by the Framingham Heart Study ; furthermore, no data have been provided for variability in other activation-restricted signaling pathways. Recently, approaches have been developed that adapt nonbiased candidate gene lists for more robust analyses incorporating genetic expression studies with genotyping as a more extensive approach to define the genetic basis of platelet phenotypic variability. These approaches are generally referred to as systems biology.
Etiology and pathogenesis: molecular genetics of platelet function
Platelet Molecular Machinery
Platelets contain as little as 2 × 10 −3 fg mRNA/cell (approximately 3–4 logs less RNA than a typical nucleated cell ), although younger platelets contain relatively larger amounts of mRNA. Platelets contain rough endoplasmic reticulum and polyribosomes, and retain the ability for protein biosynthesis from cytoplasmic mRNA. Quiescent platelets generally display minimal translational activity, although newly formed platelets synthesize various α-granule and membrane glycoproteins such as GPIb and GPIIb/IIIa (α IIb β 3 ). Historically the platelet mRNA content was considered static and invariant, although recent evidence has been presented for signal-dependent pre-mRNA splicing in platelets, suggesting a model whereby activation-dependent fluctuations of the mRNA pool could result in a dynamically altered platelet proteome. Furthermore, stimulation of quiescent platelets by agonists such as α-thrombin increases protein synthesis of various platelet proteins including the regulatory protein Bcl-3, the proinflammatory cytokine interleukin-1β, and the clot stabilizer plasminogen activator inhibitor 1. These data provide an evolutionary dynamism consistent with an adaptability to modify the genetic and proteomic composition in real time. This novel paradigmatic shift reinforces the need to comprehensively dissect the structural genomic/proteomic components to fully appreciate the dynamic nature of platelet responses in normal and diseased conditions.
Laboratory features: integrated systems approaches to dissect platelet function
Theoretically, the integration of informational databases provides for opportunities to comprehensively analyze platelet function. Utilization of genetic, functional, and proteomic datasets provides a highly innovative dissection of the entire platelet functional repertoire within a systems biology framework, an approach considerably broader (with greater implications) than simple analyses of limited differences identified using transcriptomic or proteomic profiling strategies.
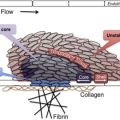
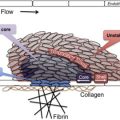
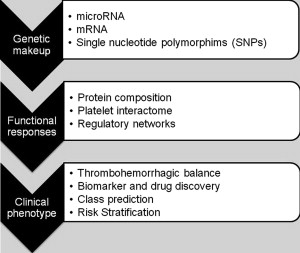
As shown in Fig. 1 , comprehensive systems biology should take into account genetic and proteomic datasets to iteratively analyze platelet function as a predictor of clinical phenotype (referred to as the platelet thrombohemorrhagic balance). Extensive efforts have gone into applying state-of-the-art technologies to comprehensively define the genetic and protein composition of platelets in normal and diseased states. This information provides the foundation for integrated analytical studies relevant to platelet function and thrombosis risk. The availability of miRNA, genotypic, and proteomic data provide unique opportunities to dissect integrated networks regulating platelet function, information that can be extracted using powerful resources currently being assembled by the platelet research community. Details of some of these databases are provided in Table 1 .
| Name | Web Site | Information |
|---|---|---|
| Swiss-PROT | http://ca.expasy.org | Platelet proteome on a 2-dimensional gel |
| Reactome | http://www.reactome.org | Functional platelet complex pathways |
| Human Protein Reference Database | http://www.hprd.org | PhosphoMotif Finder |
| PlateletWeb | http://plateletweb.bioapps.biozentrum.uni-wuerzburg.de | Centralized resource for platelet proteome, interactome, and phosphorylation |
| HaemAtlas | http://t1dbase.org/page/HaemAtlasHome | mRNA expression profiles of hematopoietic cells |
Outlined here are the various technological approaches that have provided the framework for comprehensive approaches designed to gradually gather platelet function into a clinically relevant framework. Note that the information provided is presented as an overview and is not intended to be comprehensive in scope.
Transcriptomic Analyses Focusing on Platelet mRNAs
Development of global platelet transcript profiling technologies such as microarray and serial analysis of gene expression (SAGE) in conjunction with completion of the Human Genome project led to understanding of the complex processes of gene interactions within a living cell. Unlike microarray, SAGE represents an “open transcript profiling system,” which can detect any transcript. “Classical” SAGE relies on the observation that short (<10 bp) sequences (“tags”) within 3′-mRNAs can stringently discriminate among the approximately 30,000 genes comprising the human genome, assuming a random nucleotide distribution along a 9-bp stretch (4 9 bp = 262,244 random nucleotide combinations). The sequence of each tag, along with its positional location, uniquely identifies the gene from which it is derived, and differentially expressed genes can be identified in a quantitative manner because the frequency of tag detection reflects the steady-state mRNA level of the cellular transcriptome ( www.sagenet.org ). With relatively deep sampling (>50,000 tags), genes expressed at low levels (<0.01% of total mRNA) can be readily identified, although reliable identification of splice-variant forms still represents a scientific challenge. Several modifications to the original protocols have been devised for (1) generation of longer tags as a means of providing more definitive “tag-to-gene” identification, (2) efficient identification of low-abundant transcripts using subtractive SAGE techniques, and (3) amplification techniques to circumvent small mRNA starting material.
Initial successful characterization of platelet-derived mRNA transcripts have been achieved using construction of platelet-specific cDNA libraries and single-gene polymerase chain reaction (PCR) technology. A limited number of published microarray studies using platelet-derived mRNAs have been described, with generally concordant agreements on transcript quantitation and gene-expression patterns. Furthermore, it has become clear that this approach provides an efficient means to identify novel genes and proteins functionally expressed in human platelets. Not surprisingly, platelets retain fewer transcripts than those found in nucleated cells, ranging from approximately 1600 to 3000 mRNAs. The small number of platelet-expressed transcripts reflects the lack of ongoing transcription in the anucleate platelet.
The combination of the complementary transcript profiling techniques microarray and SAGE allowed validation of most abundant platelet transcripts. Initial studies of the limited platelet SAGE library (2033 tags) demonstrated that 89% of platelet RNA tags are mitochondrial (mt) transcripts, presumably related to persistent mt-transcription in the absence of nuclear-derived transcripts. Microarray alone does not detect mitochondrial transcripts because specific probes for human mitochondrion are generally not present on the microarray chip. Recently, analysis of a more comprehensive platelet SAGE library (25,000 tags) revealed that approximately 50% (12,609 tags) of platelet SAGE tags are nucleus derived, whereas the remaining 50% are of mitochondrial origin. However, the overrepresentation of mitochondrial transcripts in platelets is considerably greater than that of its closest cell type, skeletal muscle, in which mitochondrial SAGE tags constitute 20% to 25%.
Platelet MicroRNA Transcriptomic Studies
miRNAs are a special group of short RNA species consisting of 21 to 24 nucleotides, and known to interact with target mRNAs to affect their translational efficacy. miRNA regulation represents an important posttranscriptional control to the primary cellular regulatory pathway through which gene expression defines final protein synthesis. Recent data have demonstrated that both megakaryocytes and platelets retain an abundant and diverse array of miRNAs, and have suggested an extensive regulatory role of hematopoietic-specific miRNAs in the megakaryocytic proliferation and differentiation process. Examples include interaction between miR-10a and its direct target HOXA-1 , miR-150’s role in the differentiation process of megakaryocyte-erythrocyte progenitor cells, and miR-130’s repression of transcription factor MAFB . Not surprisingly, dysregulated miRNA expression patterns have been described in myeloproliferative disorders, further implicating discrete miRNAs in lineage commitment during normal or dysregulated hematopoiesis.
In addition to precursor megakaryocytes, platelets also retain a competent miRNA pathway capable of converting precursor miRNAs through functional Dicer/Argonaute 2 (Ago2) complexes. Furthermore, evidence has been provided that Ago2–miRNA-223 complexes specifically regulate expression of the functionally important platelet purinergic P2Y 12 ADP receptor. Additional data suggest that miRNAs (miR-28) can modulate expression of the c- mpl thrombopoietin platelet receptor and that miR-96-mediated regulation of endobrevin/VAMP8 (vesicle-associated membrane protein 8) affects human platelet functional responsiveness. The potential importance of a functionally competent miRNA pathway is underscored by the unusually high miRNA/mRNA platelet ratios in comparison with other hematopoietic cells such as granulocytes and megakaryocytes, suggesting expanded layers for posttranscriptional regulation in platelets.
Platelet Proteomic Studies
Platelet proteomic studies can be grouped into distinct yet overlapping subcategories: proteomic analyses of quiescent platelets (the static platelet proteome) or of activated platelets. In toto proteomic strategies identified many platelet proteins, although subsequent approaches to dissect the changes that occur with platelet activation subcategorized platelet fractions (ie, membrane proteins) or functional end points (ie, phosphorylation patterns, microparticles, and so forth) in response to external stimuli. Such “subproteome” studies use the same tandem mass-spectroscopic technologies, but provide a more detailed analysis of function over time.
Quiescent platelets
Initial attempts to analyze the platelet proteome focused on characterizing proteins in resting platelets using 2-dimensional gel electrophoresis (2-DE) and in-gel protein detection using monoclonal antibodies. Similar techniques have been applied to characterize tyrosine-phosphorylated proteins in resting platelets and to establish a platelet protein map. More detailed profiling of platelet proteins using focused isoelectric gradients (pI range 5–11) identified 760 protein features corresponding to 311 different genes, resulting in the annotation of 54% of the 2-DE proteome map. Newer techniques include combined fractional diagonal chromatography (COFRADIC), a non–gel-based technique whereby peptide sets are sorted in a diagonal reverse-phase chromatographic system through a specific modification of their side chains. Modifications of this technology identified a core set of 641 platelet proteins, and classification using Gene Ontology demonstrated that 16% were membrane proteins, and 64% were classified as members of cytoskeleton, endoplasmic reticulum, mitochondria, cytosol, or Golgi apparatus. Somewhat unexpectedly, nearly 20% were classified as nucleus restricted; because platelets are anucleate, this observation would suggest that these proteins are megakaryocyte remnants.
Membrane proteins
Progressive, highly sophisticated proteomic methodologies have been applied to study platelet membrane receptors. In general, the study of platelet integral membrane proteins and surface receptors using 2-DE has limitations: (1) the low solubility of these proteins, (2) their association with the platelet membrane, (3) their high molecular weight, and (4) the presence of highly abundant cytoskeleton actin. By applying COFRADIC technologies, studies predicted the presence of 87 putative helix-spanning membrane proteins. A more focused analysis was pursued by enriching membrane proteins before protein identification using microcapillary liquid chromatography systems coupled to tandem mass spectrometry (referred to as μLC-MS/MS). Two distinct solubilization methods were used to reduce the overrepresentation of cytoskeleton proteins, providing for identification of 233 established or putative transmembrane proteins. A more integrated approach combining microarray analysis and mass-spectrometric techniques has been used to identify novel membrane proteins that signal during platelet aggregation, and to characterize tyrosine, threonine, or serine phosphorylated residues on platelet aggregation. Two-phase partitioning and multidimensional protein identification technology (MudPIT) resulted in an assembled dataset of 1282 proteins.
The platelet secretome and its granules
Various proteomic strategies have identified protein subsets that are secreted during platelet activation. Initial studies identified 82 secreted proteins, although a subsequent more robust analysis using thrombin-stimulated platelets expanded this list to more than 300 secreted proteins. Nearly 28% were not known to be released by any cell type, and one-third of the platelet-secreted proteins were previously known. Several of the secreted proteins have previously been described in human atherosclerotic lesions, but are absent in normal vasculature; this approach therefore has the potential to identify putative future targets for drug development in modulating atherosclerosis. Similar approaches have been applied for comprehensive profiling of subfractionated α-granules and dense platelet granules.
Posttranslational studies: the platelet phosphoproteome and glycoproteome
Initial studies to dissect signaling events downstream of thrombin activation used phosphotyrosine antibodies coupled to various proteomic detection technologies including 2-DE. By using the thrombin receptor activating peptide to specifically activate proteinase activated receptor 1, 62 differentially phosphorylated proteins were detected, 41 of which were identified by μLC-MS/MS. Of note, 8 of these appeared to be novel proteins and/or modifications, and the protein repertoire was shown to originate from 31 genes, further highlighting how alternative splicing expands the platelet proteome. Similarly, a novel approach for enrichment of sialylated glycoproteins resulted in the separation of a large number of glycopeptides from the bulk of nonmodified platelet peptides. Recently applied technical advances using phosphopeptide enrichment will likely enhance our understanding of these signal-dependent activation responses.
Platelet microparticles
Platelet activation in vivo causes release of 2 distinct membrane vesicles: microparticles (which bud from the plasma membrane) and exosomes. Microparticles range in size between 0.1 and 1.0 μm, whereas exosomes are smaller (range 40–100 nm). Platelet microparticles retain procoagulant activity, and play hemostatically critical roles in several clinical disorders including heparin-induced thrombocytopenia and immune thrombocytopenic purpura. A recent proteomic study identified 578 proteins that contain this subcellular proteome, 380 of which had not been previously described in platelet proteomic studies, suggesting these platelet fragments have a unique protein composition.
Clinical analyses focused on health and disease
Platelet Transcriptomics: Essential Thrombocythemia as a Paradigm
Direct analysis of platelet mRNAs has been applied to identify transcriptomic differences between normal platelets and those of patients with essential thrombocythemia (ET). ET represents a myeloproliferative disorder subtype characterized by increased proliferation of megakaryocytes, resulting in elevated circulating platelets. Initial studies designed to establish a proof of principle analyzed platelets from 6 ET patients and 5 healthy controls. Apheresis platelets were used and were purified and analyzed using the Affymetrix HU133A microarray chip, which contains probe sets for 22,283 transcripts. Computational analysis demonstrated distinctly different molecular signatures of normal and ET platelets, and signatures that clearly cosegregated from leukocyte fractions studied in parallel. Collectively, ET platelets demonstrated greater numbers of expressed transcripts in comparison with normal controls, although considerably less than the transcript numbers generally found in nucleated cells and leukocyte fractions. Stringent analyses (ie, marginal or present in all of the arrays within a single group) established the presence of 1840 transcripts expressed in ET platelets versus 1086 transcripts expressed in platelets from healthy controls ( P <.03). An unsupervised, hierarchical clustering analysis demonstrated that all ET platelet samples were grouped together on the basis of gene-expression similarities, with only 1 normal sample misclassified as ET. Comparison of leukocyte and platelet transcript profiles allowed delineation of “platelet-restricted genes”; that is, identification of genes whose expression was restricted to platelets (n = 126), an observation that provided unique opportunities to develop platelet class prediction models using a uniquely designed platelet gene chip (see later discussion).
Using a one-way analysis of variance, 170 genes were identified as differentially expressed between normal and ET platelets, most of which were upregulated in ET platelets. Only 29 genes were downregulated in ET platelets, of which a single platelet-restricted gene ( HSD17B3 ) was expressed in all normal platelet arrays, and uniquely underexpressed in ET compared with normal platelets. The HSD17B3 gene encoding the type 3 17β-hydroxysteroid dehydrogenase (17β-HSD3) belongs to the large family of steroid dehydrogenases, and encodes an enzyme that catalyzes conversion of 4-androstenedione to testosterone. Of interest, this enzyme is regarded as testis specific, and molecular defects of the HSD17B3 gene are causally implicated in male pseudohermaphroditism. These data provided the first evidence that HSD17B3 and HSD17B12 are expressed in human platelets and may be involved in ET. Functional 17βHSD3 activity studies demonstrated that platelets retain the capacity to convert testosterone to 4-androstenedione. Furthermore, the high level of expression of HSD17B12 transcript in ET platelets was unassociated with overall enzymatic capabilities in androgen biosynthesis. Collectively, these data provided the first evidence that genome-wide platelet transcript profiling can be adapted to study molecular signatures of platelet-associated diseases. Evidence that platelets retain 17β-HSD3 activity, express distinct subtypes of 17β-HSD enzymes, and demonstrate altered HSD17B expression patterns in a disorder known to be associated with thrombohemorrhagic risk provided novel insights into the interplay between sex hormones, platelet function, and vascular diseases.
Platelet MicroRNA Studies in Human Diseases
Several recent studies have focused on miRNA differences in human hematopoietic stem cell diseases, although studies in human platelets are more limited. Emerging evidence has implicated miRNAs in the control of megakaryocytopoiesis and in progenitor fate during the megakaryocyte-erythroid transition, presumably by modulating expression of key transcriptional regulators. Furthermore, distinct patterns of miRNA expression have been seen in differentiated hematopoietic cells and in subsets of patients with myeloproliferative neoplasms, further implicating discrete miRNAs in lineage commitment during normal or dysregulated hematopoiesis. To directly extend these studies to human platelets, research from the author’s laboratory has analyzed miRNA expression patterns in clinical situations of enhanced platelet production, comparing patterns in ET with those of patients with reactive thrombocytosis (RT). Of note, a nearly identical, unique 21-miRNA signature associated with both conditions was identified. The majority of the differentially expressed miRNAs displayed congruent directional behavior irrespective of the thrombocytosis etiology, supporting a molecular model whereby normal and exaggerated thrombopoiesis represent genetically defined continuums along a convergent pathway, rather than qualitatively distinct genetic entities. Thus, subjects with ET display the most exaggerated expression patterns, whereas those with RT represent an intermediary subset, collectively defined by quantitatively distinct expression patterns of the 21-miRNA signature. These data reinforce and extend to miRNAs the paradigm that quantitative differences involving the genetic fingerprint may be associated with (or confer) distinct phenotypes, that is, a self-limiting phenotype seen in RT rather than the sustained phenotype associated with ET. Given the convergence of c-mpl /Tpo and Jak/Stat pathways in normal and myeloproliferative-associated platelet production, it is not unexpected that the 21-miRNA thrombocytosis expression signatures remained concordant, the notable exception being miR 144/144* , which was specifically upregulated in RT and downregulated in ET. It is interesting that miR 144 is known to be transcribed with miR 451 on a single precursor RNA (pri-miRNA), and both are lineage (erythroid)-enriched in hematopoiesis models using zebrafish ( Danio rerio ). The miR 144/451 locus is a direct GATA1 transcriptional target, although effects on erythropoiesis appear to be restricted to miR 451 , with no evidence that either miRNA has an effect on thrombopoiesis. Although the orthologous miR 144/451 locus is conserved on human chromosome 17, the author saw no evidence for dysregulated miR 451 during thrombocytosis, further supporting the concept that these contiguous genes seem to have divergent effects on the megakaryocyte/erythroid transition. These data imply that distinct pathways of thrombopoiesis may be defined by the lineage-restricted expression patterns (or cognizant transcriptional regulators) of miR 144, a direction of research that remains in progress.
Nonplatelet Transcriptomic Studies Designed to Study Thrombotic Risk
The application of gene profiling has been used in clinical diseases associated with thrombosis, using either platelets or peripheral blood mononuclear cells (PBMCs) as the cellular source ( Table 2 ). Two studies have reported gene-expression changes associated with ischemic cerebrovascular stroke, differing in the use of PBMCs or neutrophils as the cellular source. Although neutrophil expression patterns appeared to represent more optimal stroke predictors, gene lists could not differentiate between ischemic and hemorrhagic stroke. Another study used leukocytes to identify 108 differentially expressed genes among subjects with CAD in comparison with healthy control subjects. These results were compared with those of gene-expression studies in murine models of atherosclerosis, and their relevance was reinforced by good correlation with human atherosclerotic lesion progression. By contrast, cross-species patterns in acute stroke correlated poorly between rats and humans. Studies focusing on cardiopulmonary bypass (CPB) demonstrated a “primed” phenotype of circulating PBMCs in which adhesion and signaling factors were overexpressed. A similar study demonstrated that the CPB coding circuit affected leukocyte gene-expression patterns, specifically demonstrating that heparin caused a more profound alteration in leukocyte gene expression when compared with a nonheparin protein-coating biomaterial. The antiphospholipid antibody syndrome is a strong risk factor for arterial thrombosis, and a recent study demonstrated that PBMC gene-expression patterns predicted an individual’s predisposition to developing thrombosis. No attempt was made to subfractionate the starting mononuclear cell population, which also included platelets. This study was limited by its retrospective design, and some of the differences could have been secondary to the primary thrombotic event.