thresholds for activation and abrogate activation responses to terminate an immune reaction.27
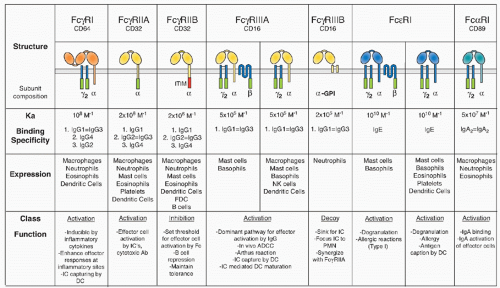
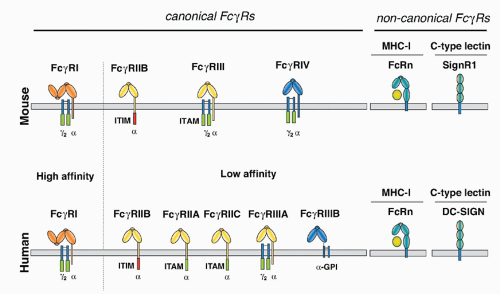
of differentiation (CD)20 family of tetraspan molecules.24 An ITAM sequence is found in the intracytoplasmic carboxy domain. In mast cells and basophils, the β chain assembles into an αβγ2 complex with the α chain belonging to either FcγRIII or FcεRI. Its presence is required for assembly of FcεRI in rodents. In humans, however, αγ2 complexes of FcεRI are found in monocytes, Langerhans cells, and dendritic cells (DCs), in addition to the αβγ2 complexes found in mast cells and basophils. The ITAM motif found in the β subunit is not an autonomous activation sequence but functions as a signaling amplifier of the ITAM found in the γsubunits.40
and trastuzumab in mouse models, demonstrating an important role of cellular FcγRs for the activity of these therapeutic antibodies in vivo.54,55 Four amino acids are polymorphic for FcγRIIIB at positions 18, 47, 64, and 88, which contribute to the neutrophil antigen polymorphisms for this receptor. Several studies have attempted to link specific FcR polymorphisms or copy number variations to autoimmune diseases, specifically to systemic lupus erythematosus (SLE).56 Recent studies have reported associations in susceptibility to SLE in both murine and human populations with levels of FcγRIIB
expression or alleles of FcγRIIB.57 Reduced expression of FcγRIIB on activated B cells, such as memory cells, has been seen in patients with SLE and chronic inflammatory demyelinating polyneuropathy (CIDP), and is associated with a promoter polymorphism.58,59 In mouse strains that develop a spontaneous, lupus-like disease, restoring the level of FcγRIIB expression on their B cells to a wild-type level reverses disease.60,61 A higher incidence of an allele of FcγRIIB has been reported in several populations with autoimmune disease. This allele, found in the transmembrane domain of the receptor, is suggested to result in a hypomorphic phenotype, similar to the reduced expression observed.57 Of note, despite increasing the risk for development of SLE, this FcγRIIB allele seems to decrease the likelihood of malaria infections and the severity of disease and can be found at increased frequencies in areas of the world where malaria is endemic.62,63
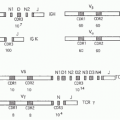
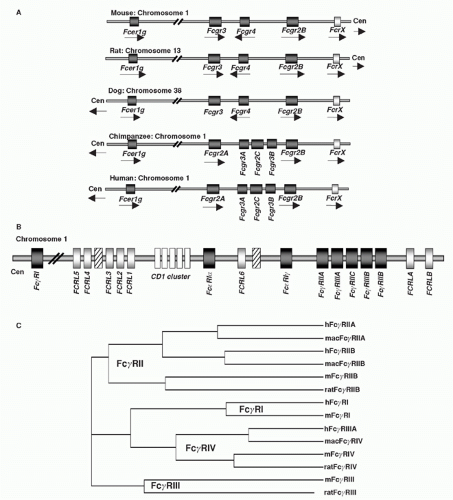 FIG. 24.2. Chromosomal Organization of Fc Receptor (FcR) and FcR-Like Genes. A: Localization of FcR genes in different species. B: The human FcR locus on chromosome 1. Classical FcR genes are shown in dark grey, FCRL genes as light grey, and the CD1 gene cluster on chromosome 1 as white boxes. Pseudogenes are indicated as hatched boxes. (adapted from Davis et al.44). C: The cladogramm shows the alignment of selective classical FcRs of humans (h), macaques (mac), mice (m), and rats. |
in mice, precluding the expression of the αγ2 complex.24 In humans, this form of the receptor is widely expressed on monocytes, Langerhans cells, and DCs, and is likely to be found on mast cells and basophils as well. This difference in FcεRI subunit composition is likely to result in functional differences as well. Although these specific interspecies differences are important, the fundamental organization of the canonical FcR system, with activation and inhibitory signaling through a shared ligand specificity coupled to opposing signaling pathways, is well conserved, as is the role of the noncanonical FcRs in mediating the immunomodulatory functions of sialylated IgG. Thus, conclusions regarding the function of this expanded FcR system in immunity by the analysis of murine models are relevant to an understanding of the role of these receptors to human immunity as well.
found in IgE). Specificity is generated among the receptor-ligand pairs in a variable region connecting the two extracellular domains that is in contact with the lower hinge region of the Fc fragment (residues 234 to 238), a region not conserved among the IgGs and IgE. The binding region of FcRs to their Ig ligands does not overlap with other Fc binding molecules such as protein A, protein G, and FcRn.
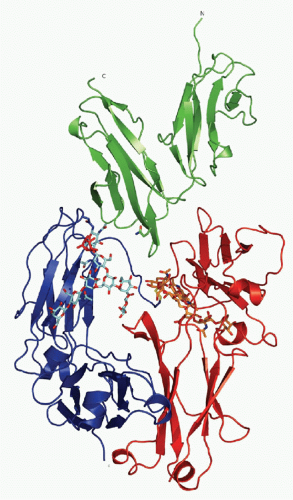 FIG. 24.4. Ribbon Diagram of Crystallizable Fragment (Fc) Receptor III-Immunoglobulin (IG)G1 Fc Structure. The carbohydrate moiety attached to both CH2 domains of the IgG Fc-fragment is shown as a stick and ball model. The extracellular domains of Fc receptor III are shown, together with the Fc fragment of IgG1. See text for details. Adapted from Sondermann et al.,15 with permission. |
been reported to influence affinity for ligand, as demonstrated for the common γchain associating with FcγRIIIA.90 Its affinity for IgG1 is higher than the GPI-anchored form of this receptor, FcγRIIIB.
TABLE 24.1 Affinities of Mouse and Human Fc Receptors to Different Antibody Isotypes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree