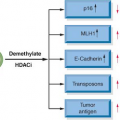
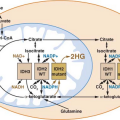
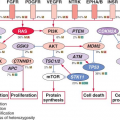
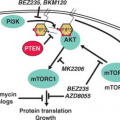
Syndrome |
OMIM Entrya |
Major Tumor Types |
Mode of Inheritance |
Genes |
HEREDITARY GASTROINTESTINAL MALIGNANCIES |
Adenomatous polyposis of the colon |
175100 |
Colon, thyroid, stomach, intestine, hepatoblastoma |
Dominant |
APC |
Juvenile polyposis |
174900 |
Gastrointestinal |
Dominant |
SMAD4/DPC4 |
Peutz-Jeghers syndrome |
175200 |
Intestinal, ovarian, pancreatic |
Dominant |
STK11 |
GENODERMATOSES WITH CANCER PREDISPOSITION |
Nevoid basal cell carcinoma syndrome |
109400 |
Skin, medulloblastoma |
Dominant |
PTCH |
Neurofibromatosis type 1 |
162200 |
Neurofibroma, optic pathway glioma, peripheral nerve sheath tumor |
Dominant |
NF1 |
Neurofibromatosis type 2 |
101000 |
Vestibular schwannoma |
Dominant |
NF2 |
Tuberous sclerosis |
191100 |
Hamartoma, renal angiomyolipoma, renal cell carcinoma |
Dominant |
TSC1/TSC2 |
Xeroderma pigmentosum |
278730, 278700,
278720, 278760,
278740, 278780,
278750, 133510 |
Skin, melanoma, leukemia |
Recessive |
XPA,B,C,D,E,F,G, POLH |
Rothmund Thomson syndrome |
268400 |
Skin, bone |
Recessive |
RECQL4 |
LEUKEMIA/LYMPHOMA PREDISPOSITION SYNDROMES |
Bloom syndrome |
210900 |
Leukemia, lymphoma, skin |
Recessive |
BLM |
Fanconi anemia |
227650 |
Leukemia, squamous cell carcinoma, gynecological system |
Recessive |
FANCA,B,C,D2,
E,F,G |
Shwachman Diamond syndrome |
260400 |
Leukemia/myelodysplasia |
Recessive |
SBDS |
Nijmegen breakage syndrome |
251260 |
Lymphoma, medulloblastoma, glioma |
Recessive |
NBS1 |
Ataxia telangiectasia |
208900 |
Leukemia, lymphoma |
Recessive |
ATM |
GENITOURINARY CANCER PREDISPOSITION SYNDROMES |
Simpson-Golabi-Behmel syndrome |
312870 |
Embryonal tumors, Wilms tumor |
X-linked |
GPC3 |
Von Hippel-Lindau syndrome |
193300 |
Retinal and central nervous hemangioblastoma, pheochromocytoma, renal cell carcinoma |
Dominant |
VHL |
Beckwith-Wiedemann syndrome |
130650 |
Wilms tumor, hepatoblastoma, adrenal carcinoma, rhabdomyosarcoma |
Dominant |
CDKN1C/NSD1 |
Wilms tumor syndrome |
194070 |
Wilms tumor |
Dominant |
WT1 |
WAGR syndrome |
194072 |
Wilms tumor, gonadoblastoma |
Dominant |
WT1 |
Costello syndrome |
218040 |
Neuroblastoma, rhabdomyosarcoma, bladder carcinoma |
Dominant |
H-Ras |
CENTRAL NERVOUS SYSTEM PREDISPOSITION SYNDROMES |
Retinoblastoma |
180200 |
Retinoblastoma, osteosarcoma |
Dominant |
RB1 |
Rhabdoid predisposition syndrome |
601607 |
Rhabdoid tumor, medulloblastoma, choroid plexus tumor |
|
SNF5/INI1 |
Medulloblastoma predisposition |
607035 |
Medulloblastoma |
Dominant |
SUFU |
SARCOMA/BONE CANCER PREDISPOSITION SYNDROMES |
Li-Fraumeni syndrome |
151623 |
Soft tissue sarcoma, osteosarcoma, breast, adrenocortical carcinoma, leukemia, brain tumor |
Dominant |
TP53 |
Multiple exostosis |
133700, 133701 |
Chondrosarcoma |
Dominant |
EXT1/EXT2 |
Werner syndrome |
277700 |
Osteosarcoma, meningioma |
Recessive |
WRN |
ENDOCRINE CANCER PREDISPOSITION SYNDROMES |
MEN1 |
131000 |
Pancreatic islet cell tumor, pituitary adenoma, parathyroid adenoma |
Dominant |
MEN1 |
MEN2 |
171400 |
Medullary thyroid carcinoma, pheochromocytoma, parathyroid hyperplasia |
Dominant |
RET |
WAGR, Wilms tumor, aniridia, genitourinary abnormalities, mental retardation; MEN, multiple endocrine neoplasia. |
a Online Mendelian Inheritance in Man, http://www-ncbi-nlm-nih-gov.proxy3.library.mcgill.ca/Omim/getmorbid.cgi. (Adapted from ref. 137.) |
|