Sites of B Lymphopoiesis during Ontogeny
In the mouse, hematopoiesis occurs predominantly in the fetal liver prior to birth, in the spleen just prior to and shortly after birth, and in the bone marrow thereafter. Prior to liver hematopoiesis, the blood islands of the yolk sac (YS) contain the first identifiable hematopoietic cells, nucleated erythrocytes with embryonic forms of hemoglobin.
1 However, these early YS precursors appear incapable of generating other blood cell lineages, and generation of all blood cell types, including lymphocytes,
2,3 initiates at around 9 to 10 dpc in an embryonic region referred to as the splanchnopleura/AGM (or simply Sp/AGM). Cells from this site are capable of longterm repopulation of lethally irradiated adult recipients with all blood lineages.
4,5 These cells colonize the fetal liver at about 11 dpc, initiating hematopoiesis there. Thus, there are two sites of very early hematopoietic precursors, with one in the YS largely limited to erythropoiesis and the other in the Sp/AGM capable of complete (referred to as “definitive”) hematopoiesis. However, it may be that precursors in the YS have a broader lineage potential in the fetal microenvironment, as when they are injected directly into the newborn liver.
6 HSCs capable of developing into all the blood cell types are produced in the Sp/AGM and migrate to the fetal liver at about d10. Thereafter, B-lineage cells develop largely in a wave, with earlier stages present at earlier times and later stage predominating at later times, close to (and shortly after) birth.
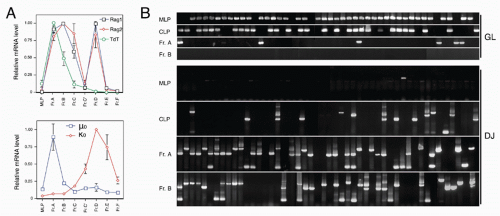
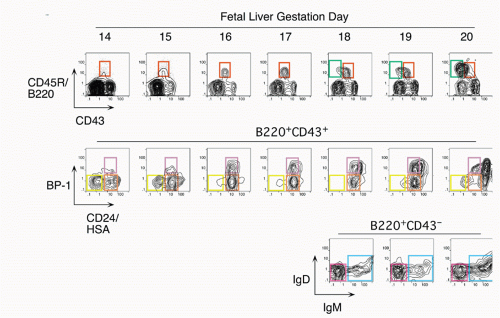
7,8 This progression with gestation day is easily visualized by staining with antibodies that delineate B-cell
development, as shown in
Figure 8.2. Early precursors can also be found in the fetal omentum.
9 In contrast with the bone marrow, cells at most differentiation stages in the fetal liver appear to be rapidly proliferating so that larger and larger numbers of B-lineage cells are detected at progressive days of gestation. Another distinction of fetal liver from bone marrow development in the adult is the absence of terminal deoxynucleotidyl transferase (TdT),
10,11 an enzyme that mediates nontemplated addition of nucleotides at the D-J and V-D junctions of Ig heavy chain.
12,13,14 Therefore, heavy chains produced during fetal development have little or no N-region addition, and CDR3 diversity is constrained even further by favoring of short stretches of homology at the V-D and D-J junctions.
15,16 Rearrangement of certain V or D elements may also differ between fetal and adult development, as for example, the reported high utilization of the DFL16.1 segment in fetal liver.
17 Differential expression of genes other than TdT also distinguish B-cell development during fetal life from that in the adult including precursor lymphocyte regulated myosin light chain like PLRLC transcripts
11,18 and major histocompatibility complex (MHC) class II.
19,20 Interestingly, although absence of the cytokine IL-7 completely eliminates bone marrow B-lineage development,
21 it nevertheless spares some fetal development,
22 suggesting a difference in growth requirements. The B-cell progeny of this early fetal wave may largely consist of B cells quite distinct from adult-derived cells, populating the B-1 subset.
23At birth, B-cell development can also be detected in spleen, but development at this site gradually decreases to very low levels by 2 to 4 weeks of age. Over this same period, B-cell development shifts to the bone marrow and thereafter it continues for the life of the animal. B lymphopoiesis decreases in aged mice. This may be due to diminished responsiveness of precursors to IL-7.
24,25
Stem Cells, Commitment, and Early B-Cell Progenitors in Bone Marrow
B cells are continually generated from HSCs in the bone marrow of adult mice. Considerable effort has focused on evaluating the functional capacity of fractions of bone marrow cells to repopulate different lineages of blood cells; this work has progressed to the stage of defining a phenotype for such cells, with expression of c-KIT constituting an important marker in the so-called lineage negative subset.
26,27 This is the small fraction of bone marrow cells (< 5%) that lacks expression of a panel of “differentiation markers,” cell surface molecules that are expressed on later stages of various hematopoietic cell lineages. Careful analysis of this HSC fraction using additional markers has shown that it represent perhaps 1/30,000 of nucleated bone marrow cells with as few as 10 mediating multilineage repopulation in cell transfer assays.
28,29,30 An important capacity of “true” or “long-term repopulating” HSCs is their ability to give rise to cells in a recipient mouse that can also repopulate all the blood cell lineages upon retransfer into a second host, indicating a capacity for extensive self-renewal without differentiation into more restricted progenitors.
A major focus in research on hematopoiesis has been defining and characterizing lineage-restricted progenitors, such as the common myeloid and common lymphoid
progenitors (CLPs).
31,32,33 The CLP cell fraction was identified by lack of a panel of “lineage markers,” expression of the IL-7Rα chain, and distinctive, intermediate levels of c-KIT, compared to higher levels on HSCs. Initial characterization of these cells in various functional assays suggested that these cells could generate B, T, natural killer (NK), and a subset of dendritic cells but no other blood cell lineages. The reason for this restriction has been intensively studied, and downregulation of the receptor for granulocyte-myeloid colony stimulating factor has been suggested to be a key event in this process.
34 Cells with the phenotype of CLP constitute about 1/3000 of bone marrow cells. Prior to the CLP stage, multipotent progenitors exhibit low-level expression of genes characteristic of diverse cell lineages, leading to the idea that such promiscuous expression indicates chromatin accessibility that facilitates flexibility in cell fate decisions.
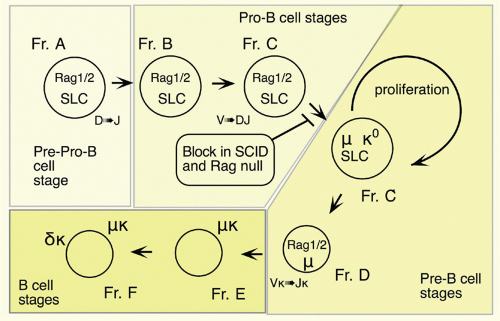
35CLP cells can give rise in short-term cultures to cells of the B lineage, naturally raising the issue of when cells become restricted to the B lineage. Most cells growing in stromal cultures give rise only to B cells upon transfer into mice, and the phenotype of these cells has been well characterized.
36 Most have at least some heavy chain rearrangement and bear the B-lineage marker CD19.
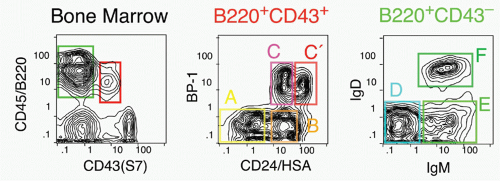
37,38 There is less certainty concerning the cells isolated directly from primary lymphoid tissues, as such cells are quite rare similar to CLPs and HSCs. Most of the CD45R/B220+ cells in bone marrow are also CD19+; such cells are committed to the production of B cells.
39 However, a subset of B220+ cells lacks detectable CD19 expression; cells within this fraction can generate CD19+ cells in short-term stromal culture with IL-7. Such cells are included in the CD43+CD24
low fraction (Fr. A, 1% of bone marrow) of B220+ cells in bone marrow, but this fraction also contains other cell types including NK-lineage precursors.
38,40 Thus, it is necessary to exclude cells lacking AA4.1 (about half
38) and expressing Ly6c.
41 Many of these Ly6c+ cells also express CD4; recent work suggests that these are plasmacytoid dendritic cells.
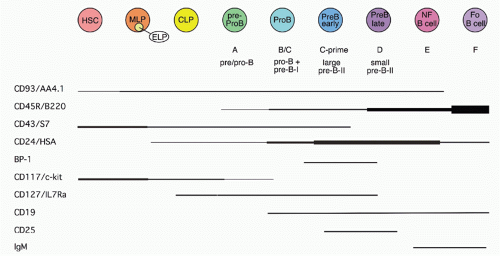
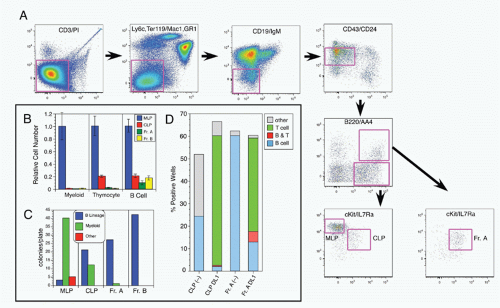
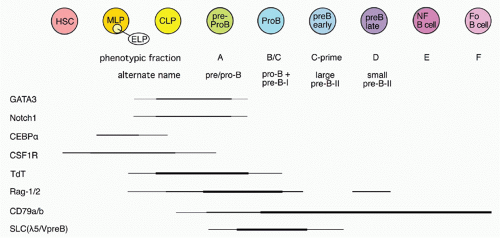
42,43 A phenotypic approach for enriching and fractionating very early B-lineage subsets is shown in
Figure 8.3A.
Careful analysis of the LIN− (including CD19) IL7Rα+cKIT+ CD45/B220− (CLP) and CD45/B220+ (Fr. A), as delineated in
Figure 8.3, modified our understanding of the earliest stages of B-cell development in bone marrow.
44 First, while CLP stage cells fail to efficiently generate myeloid cells upon transfer into irradiated hosts, they nevertheless retain significant capacity to produce such cells in short-term cultures, likely due to continued (albeit reduced) expression of receptors for myeloid growth factors. In contrast, this myeloid capacity is greatly reduced as cells begin to express CD45R/B220 (ie, become “Fr. A”), concomitant with reduced expression of receptors for myeloid growth factor receptors. Yet these Fr. A cells, while poorly reconstituting T cells in cell transfer assays, nevertheless retain the capacity to generate T-lineage cells in culture, mediated by engagement of Notch by its ligand DL1.
45 Thus, the potential for alternate hematopoietic lineages appears to be lost somewhat later in progression down the B-lineage pathway in mouse bone marrow than previously thought. On the other hand, it appears that initiation of Ig rearrangement is initiated earlier than some studies have indicated. Determination of the extent of germline deoxyribonucleic acid (DNA) segments lost upon D-J rearrangement, and the formation of such D-J segments in individual cells isolated by electronic cell sorting showed that 30% to 50% of cells in CLPs and more than 80% of cells in Fr. A contained a D-J rearrangement on at least one chromosome.
44 This is consistent with high-level expression of genes important in Ig rearrangement, including TdT, Rag-1, and Rag-2, in CLP
46 and Fr. A stage cells.
An emerging view of CLPs (and possibly even the earlier multilineage progenitor [MLP]) stage cells considers them to be early B-lineage precursors rather than branch points in the production of other hematopoietic cell lineages. Thus, analysis of CD4/CD8/CD3 “triple-negative” cells in thymus failed to identify cells with a surface phenotype comparable to CLPs, and mutant mice lacking CLPs in bone marrow nevertheless have relatively intact thymic development, leading these authors to suggest a distinct “early T progenitor” different from CLPs.
47 Furthermore, cells with T/myeloid potential, but lacking B-lineage capacity, have been described.
48 It seems reasonable to hypothesize that MLP, CLP, and Fr. A stage cells occupy a distinctive microenvironmental niche in bone marrow where they receive signals that guide them along the early stages of B-lineage development culminating in CD19+ pro-B cells that are irreversibly committed to becoming B cells due to expression of PAX5
39 (see following section). Interestingly, cells considered to be progressing down a lymphoid/B-lineage path can be redirected to become dendritic cells by signals through toll-like receptors (TLRs), suggesting that infection can profoundly alter early stages in hematopoietic development.
49Additional issues remain regarding the lineage restriction of cells at these early stages in B-cell development. For example, there is evidence that cells restricted to generating B and myeloid/macrophage (but not T) lineage may exist in the fetal liver
50 and even in bone marrow.
51 There is also apparently a different dependence of fetal liver B lymphopoiesis on expression of the transcription factor BSAP compared to bone marrow, as determined by analysis of PAX5 null mice.
52 Further comparison of B-cell development in fetal liver with that in bone marrow is needed to clarify this point. Finally, the precise delineation and characterization of B-cell precursors earlier than CLPs, prior to IL-7 expression, remains imprecise. It seems likely that at least some of the MLP stage cells mentioned previously are initiating a B-lineage program based on their expression of E2A, Rag-1, Rag-2, and TdT.
44 However, potential heterogeneity in this fraction needs to be assessed. Determination of Rag-1 transcriptional activity at the single cell level by a green fluorescent protein reporter, used for identification of the early lymphoid progenitor fraction,
53 may provide a key approach for such studies.
Transcription Factors Regulating B-Lineage Development
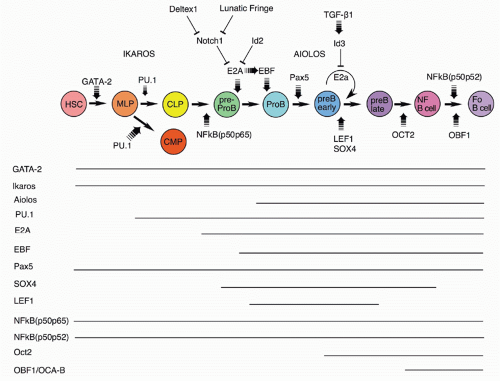
The GATA-2 and Runx1/AML1 transcription factors are required for the development of HSCs that are the precursors of all the blood cell lineages, including B cells
54,55,56,57,58 (Fig. 8.4). Experiments with the core-binding factor-associated leukemia fusion protein CBFbeta-SMMHC, whose expression
inhibits RUNX function, have revealed that its expression negatively impacts pre-pro-B-cell through pre-B-cell populations in bone marrow.
59 Although the frequency of CLP stage cells was unaffected, the expression of B-lineage associated genes (such as CD79a and λ5) was decreased, demonstrating the key importance of early RUNX activity.
Somewhat later acting, but still very early in development, is the Ikaros transcription factor.
60,61,62 Ikaros and the related transcription factor Aiolos
63 play important roles in lymphocyte development. Ikaros is expressed early in hematopoietic precursors. Ikaros null mice lack B-lineage cells
62; a different Ikaros mutant that acts as a dominant negative completely blocks lymphoid development.
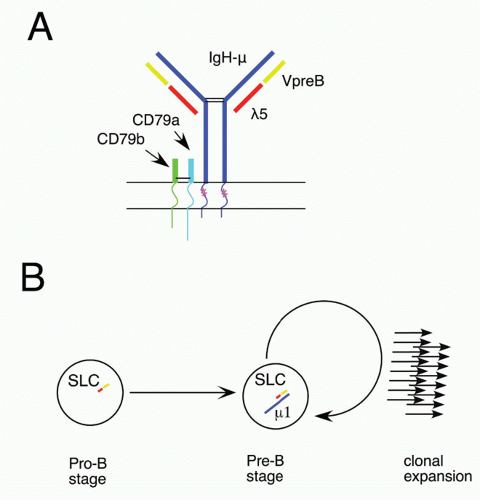
60 Ikaros activates numerous early B-lineage genes, including TdT, Rag-1, λ5, and VpreB. Expression of EBF1 in Ikaros−/− hematopoietic progenitor cells restored generation of CD19+ cells, but these cells were not committed to the B-cell fate and failed to rearrange IgH genes.
64 Thus, Ikaros acts in a transcription factor pathway, inducing EBF1 expression, and acting in concert with PAX5 to maintain B-lineage commitment, but also altering chromatin compaction around the IgH locus, which together with Rag expression results in heavy chain rearrangement. Aiolos is detected somewhat later in development at about the stage of B-lineage commitment, and its expression increases further at later stages. It is induced by pre-B-cell receptor (BCR) signaling (see section on the role of Ig heavy chain and the pre-BCR), and it acts to down-regulate transcription of λ5 (initially induced by action of Ikaros), a part of the pre-BCR complex. In this way, Ikaros family members play key roles both initiating and terminating pre-BCR signaling, a critical checkpoint in B-cell development.
The PU.1, an Ets family transcription factor, is critical for progression to the earliest stage of lymphoid development, as demonstrated by the inability of PU.1 null precursors to generate lymphocytes.
65,66 An important target of PU.1 for B-lineage development is the gene for Igβ, known as MB-1. The level of PU.1 appears to be critical for development
along the B lineage, as, while low-level expression induced in PU.1 null mice allowed B-lineage development, high-level expression blocked this and fostered myeloid lineage development,
67 likely due to differential induction of the IL-7Rα and macrophage colony-stimulating factor receptor chains.
68 In fact, retroviral mediated expression of the IL-7Rα chain complements defective B lymphopoiesis in PU.1 null bone marrow HPCs.
69 Surprisingly, recent work from several groups indicates that some B-cell development can occur in the absence of PU.1 expression.
70 Furthermore, analysis of conditional PU.1 knockout mice showed that expression of this transcription factor was not required after the pre-B-cell stage.
71E2A codes for two proteins, E12 and E47, members of the basic helix-loop-helix family of transcription factors; its induction is crucial from the earliest stages of B-lineage development, as all stages after CD19 expression are absent from E2A null mice.
72,73 These mice lack detectable D-J rearrangements, and, interestingly, such rearrangements can be induced in nonlymphoid cells by introduction of the Rag genes and ectopic expression of E2A,
74 implicating this transcription factor in the process of chromatin remodeling of the Ig heavy chain locus that permits accessibility by the recombinase machinery.
75 The regulation of E2A is crucial for B-lineage development, as negative regulators such as Notch1 and ID2 have been show to block this lineage and induce alternate cell fates, the T and NK lineages.
76,77,78,79 Consistent with this picture, ectopic expression of genes that negatively regulate Notch1, lunatic fringe, and Deltex-1 induce the B-cell fate.
80,81,82Expression of the early B-cell factor, EBF1, a member of the O/E protein transcription factor family, is requisite for progression of early B-lineage progenitors to the D-J rearranged pro-B stage (Fr. B), as shown in EBF1 null mice.
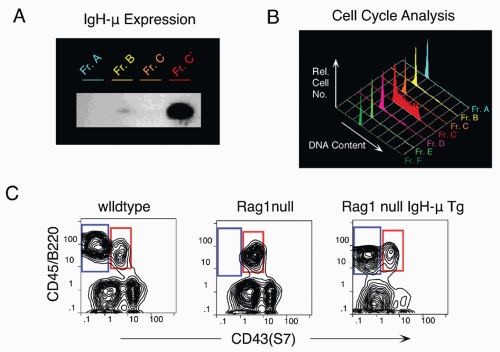
83 The expression of EBF1 is induced by action of the epigenetic histone H2A deubiquitinse MYSM1, as revealed by targeting this gene.
84 EBF1 and E2A act at a similar stage in early in B-lineage development; these two transcription factors can act together to upregulate a family of early B-lineage-specific genes, including Ig-α/β, VpreB/λ5, and Rag-1/2.
85,86 There is evidence that E2A upregulates expression of EBF1, found by transfection of E2A in a macrophage cell line,
87 suggesting an ordering of these two in development. Furthermore, recent studies showed that there are two distinct promoters for EBF1 that are regulated differently.
88 A distal promoter is activated by IL-7 signaling, E2A and EBF1, whereas a proximal promoter is regulated by PAX5, Ets1, and PU.1. Such complex regulation indicates that B-cell development occurs through the action of several feedback loops in a regulatory network that is becoming understood.
89,90BSAP, the product of the PAX5 gene, is expressed throughout B-cell development until the plasma cell stage.
91 PAX5/BSAP transcriptional targets include CD19 and BLNK; expression of this transcription factor acts to upregulate V to D-J heavy chain rearrangement.
52 Analysis of chromatin structure around the Ig heavy chain locus revealed that PAX5 induces V to D-J locus contraction, thereby promoting rearrangement.
92 PAX5 null mutant mice show an arrest in bone marrow development at the pro-B stage, likely due to the lack of complete heavy chain rearrangements and also due to the absence of the critical B-cell adaptor protein BLNK that serves to link the pre-BCR to the intracellular signaling pathway via the tyrosine kinase Syk.
93 BSAP/PAX5 also acts to repress alternate cell fates, as pro-B phenotype cells isolated from PAX5 null bone marrow can generate diverse hematopoietic cell lineages, in contrast with such cells from wild-type mice that are B-lineage restricted.
39,94 This occurs by repression of the myeloid growth factor receptor gene c-fms
95 and by repression of the Notch1 signaling pathway
96 critical for T-cell fate specification.
45,97 Conditional targeting of PAX5 in more mature B-cell stages shows that its continued expression is necessary for maintenance and function of mature B cells.
98 Finally, as mentioned previously, in contrast with bone marrow, the absence of BSAP/PAX5 arrests B-cell development prior to the B220+ stage in fetal liver, suggesting a crucial difference in the early dependence on this transcription factor.
52The Forkhead family transcription factor FoxO1 plays important roles at several stages of B-cell development.
99 Early in development, it acts to induce expression of the receptor for IL-7 at the CLP stage. It also functions to regulate Rag-1 and Rag-2 expression during heavy and light chain rearranging stages of development.
100 Finally, in mature B cells, it is needed for normal expression of L-selectin, a homing receptor important for normal recirculation of peripheral B cells through the lymphatics. GA binding protein, a ubiquitously expressed Ets family transcription factor, is another player regulating expression of the IL-7R.
101 Importantly, through interaction with PAX5, it acts in concert to induce expression of critical PAX5 target genes such as CD79a. There is recent evidence that FoxO1 regulates Ikaros activity by altering splicing of its messenger ribonucleic acid (mRNA), rather than altering Ikaros transcription.
102 FoxO1 activation of Ikaros was sufficient for induction of rearrangement of proximal VH genes, but expression of PAX5 was required for rearrangement of distal VH genes. Thus, FoxO1, Ikaros, and PAX5 appear to function in a network to coordinate the ordered rearrangement of Ig genes during B-cell development.
Lymphoid enhancer binding factor (LEF-1) shows a pattern of expression restricted to the pro-B and pre-B stages of B-cell development.
103 Targeted inactivation of the LEF-1 gene allows B-cell development but with reduced numbers.
104 This is because LEF-1 regulates transcription of the Wnt/β-catenin signaling pathway whose activation increases proliferation and decreases apoptosis of early B-lineage cells. In fact, exposure of normal pro-B cells to Wnt protein induces their proliferation.
104 Interestingly, there is a counterproliferative signal that can act at the pre-B proliferative stage, mediated by transforming growth factor-β1.
105 It appears that this occurs due to induction of the ID3 inhibitor that negatively regulates the activity of E2A.
106 Another transcription factor whose expression is similar to LEF-1 is SOX-4; its inactivation also results in the inability of normal early B-lineage cell expansion and a block at the pro-B stage.
107Several forms of NF-κb subunits are expressed throughout B-cell development; this transcription factor can regulate kappa light chain expression and also growth factor signaling.
108 Mice lacking the p65 subunit die before birth, so development must be analyzed by transfer of fetal liver precursors into wild-type recipients. Such experiments showed diminished B-lineage cell numbers, but the major defect was in mature B-cell mitogenic responses.
109 Mice lacking the p50 subunit showed relatively normal B-cell development but again poor response to mitogen by mature B cells.
109 However, mice lacking both the p50 and p65 subunits failed to generate any B220+ B-lineage cells. Curiously, when mixed with wild-type fetal liver cells, normal numbers of mature B cells could be generated from the double-defective precursors, suggesting that the defect could be overcome by secreted or membrane-bound signals provided by the wild-type precursors. Another double mutant, p50p52, showed a late stage defect in B-cell development, with a failure to generate mature B cells in spleen.
110Inactivation of the Oct-2 transcription factor results in neonatal lethality, but transfers of fetal precursors can reconstitute lymphoid cells in wild-type recipients, allowing assessment of effects on the B lineage. Such studies have shown that fewer mature follicular B cells are generated in these mice and B-1 (CD5+) B cells are completely eliminated.
111,112,113 Similarly, the Oct binding factor, OBF-1, also known as OCA-B and BOB-1, appears to function in the maturation of newly formed B cells in the bone marrow to become follicular B cells in the periphery, as inactivation of this gene resulted in a significant deficit in mature B cells.
114,115,116 Both of these transcription factors have been shown to regulate the follicular B-cell chemokine receptor CXCR5, which may explain at least part of the defect.
117 Curiously, unlike Oct-2 null mice, there was reportedly no deficit in B-1 B cells in OBF-1 null mice. Interestingly, when the OBF-1 mutant mouse is crossed with btk-deficient mice, there is a complete lack of peripheral B-cell generation,
118 suggesting that this transcription factor may function in the BCR-mediated selection of mature B cells.
Impact of Microribonucleic Acids on B-Cell Development
The miRNAs are small noncoding ribonucleic acids that facilitate the degradation of mRNAs and thereby act at a posttranscriptional level to regulate gene expression. The generation of mature functional miRNAs requires action of Dicer, a protein that cleaves pre-miRNAs, so the targeting of Dicer allows assessment of the global effect of miRNA on B-cell development. Ablation of Dicer in early B-lineage progenitors results in a block at the pro- to pre-B cell stage, likely due to upregulation of the proapoptotic molecule Bim.
119 Counteraction of Bim function by a Bcl-2 transgene reveal further dysregulation of normal development, including nontemplated nucleotide addition (N-regions) at the Ig light chain V-J junction, due to aberrant expression of the terminal deoxynucleotidyl transferase gene that is normally extinguished at the pre-B-cell stage.
Another approach for assessing the importance of specific miRNAs is direct overexpression or knockdown of expression; such a study with miR-150 reveal its role in regulating c-Myb, a transcription factor that regulates pro-B to pre-B progression and also the survival of mature B cells.
120 Transgenic overexpression of miR-17-92, a miRNA often found to be amplified in lymphoma, resulted in a lympho-proliferative syndrome and autoimmune disease, resulting in premature death.
121 One target of miR-17-92 is Bim; its decrease in the transgenic animals may have resulted in excessive cell survival and a loss of normal tolerance to self-antigens. The potential relevance to normal growth regulation is quite interesting, considering the amplification of this miRNA in some lymphomas. Another miRNA with a cancer association, miR-21, has been studied as a transgene in mice, where it results in tumors with a pre-B malignant lymphoid-like phenotype.
122 The miRNAs that are amplified in cancers and likely contribute to the neoplastic process are now termed “oncomirs.”
miRNAs can also influence lineage choice or act at key checkpoints during hematopoietic development. Retroviral provision of miR-34a in bone marrow hematopoietic progenitors blocked B-cell development at the pro-B to pre-B checkpoint, resulting in reduced numbers of mature B cells in mice repopulated with such precursors. A possible explanation for the developmental block is action of miR-34a on Foxp1, a transcription factor that appears to be important at this stage, as cotransfection of FoxO1 lacking its normal 3′ UTR target of miR-34a restored B-cell development. A novel regulator of lineage choice appears to be Let-7, a family of miRNAs regulated by the highly conserved ribonucleic acid-binding protein Lin28.
123 While Lin28 has been studied for its role in pluripotency, developmental timing, and oncogenesis, a recent study indicates that it may regulate a developmental switch in both B and T cells, such that its expression in adult hematopoietic progenitors results in reprograming development toward a more “innate-like” pattern, normally only seen during fetal/neonatal timing.
124