APPROACHES TO THE QUANTITATIVE ASSESSMENT OF GENE EXPRESSION
TRANSCRIPTION ASSAYS
Nuclear Run-On Assays.
Several assays are available that provide an index of relative rates of gene transcription (Fig. 2-2). A simple, straightforward assay is the nuclear run-on assay in which nuclei are isolated from tissue culture cells and nascent RNA chains are allowed to continue to polymerize in the presence of radiolabeled deoxyribonucleotides in vitro. This assay has the advantage that it surveys the density of nascent transcripts made from the endogenous genes of cells and, on average, is a good measure of gene transcription rates in response to the existing environmental conditions in which the cultured cells are maintained. Newly synthesized RNA is applied (hybridized) to a nylon membrane on which a cDNA target complementary to the desired RNA has been adsorbed. Radiolabeled RNA hybridized to the cDNA is determined in a radiation counter.
Cell-Free In Vitro Systems.
Rates of RNA synthesis can also be determined in broken cell or cell-free lysates to assess the relative strengths of different promoters. To restrict the newly synthesized radiolabeled RNA to a single size and, thus, to enable more ready detection by electrophoresis, a DNA template is used that does not contain guanine bases, called a G-free cassette. RNA synthesis is carried out in the absence of the guanine nucleotide. After synthesis of a specified length of RNA at the end of which guanine bases are encountered, RNA synthesis is terminated.
Transfection of Promoter-Reporters in In Vivo Cell Culture.
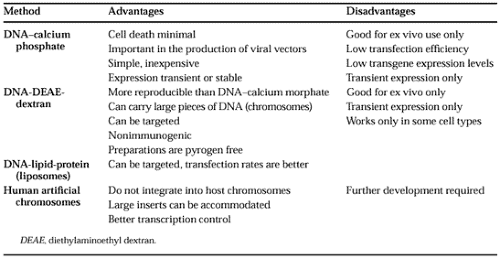
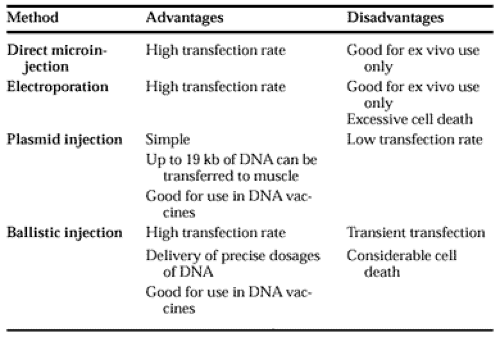
Many of the currently used assays of gene transcription employ promoter sequences fused to genes encoding proteins that can be quantitated by bioassays (e.g., bacterial chloramphenicol acetyl transferase, firefly luciferase, alkaline phosphatase, or green fluorescent protein). The hybrid DNAs, so called promoter-reporter DNAs, are introduced into tissue culture cells by one of several chemical methods (i.e., DNA adsorbed to calcium phosphate precipitates, diethylaminoethyl (DEAE)-dextran incorporated into liposomes, or human artificial chromosomes [Table 2-2]); or physical methods (i.e., electroporation, direct microinjection of DNA, or ballistic injection using a gene gun [Table 2-3]). After introduction of the reporter DNA into the cells, the transfected cells are incubated for a specified time under the desired experimental conditions, the cells are harvested, and extracts are prepared for assays of the reporter-specific enzymatic activity. By these transfection methods, cell-type specificity for the expression of genepromoter sequences can be determined by comparing promoter-reporter efficiencies in cells of different phenotypes. In addition, important transcriptional control sequences in the promoter can be mapped by DNA mutagenesis studies.
Transfection of Transcription Factor Expression Vectors.
An extension of the promoter-reporter transfection approach is to cotransfect recombinant expression plasmids encoding transcription factors that bind to control sequences in the promoter DNA and activate transcription of the reporter. By this approach, critical functional components of transcription factors and critical bases in DNA control sequences can be examined experimentally.
Transgenic In Vivo Mouse Models.
A method developed for examining specificity of tissue expression and efficiency of expression of promoter-reporter genes is their introduction into mice in vivo, using transgenic technology (see the section Genetic Manipulations in Animals In Vivo). Recombinant promoter-reporter genes are injected into the pronucleus of fertilized mouse ova and implanted into surrogate females. The tissues of transgenic neonatal mice are examined for the tissue distribution and relative
strength of the expression of the reporter function. Commonly used reporter functions are the genes encoding either β-galactosidase or green fluorescent protein.
strength of the expression of the reporter function. Commonly used reporter functions are the genes encoding either β-galactosidase or green fluorescent protein.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree