Antibody Diversity
Learning Objectives
• Recognize the three theories that explain antibody diversity
• Compare and contrast genes that code for light chains and heavy chains
• Recognize the key features of combinatorial diversity
• Explain the biologic function of complementarity regions 1, 2, and 3
• Discuss the role of the recognition signal sequence (RSS) in recombination
• Relate the biologic significance of the 12/23 rule in the recombination of V(D)J genes
• Understand the role of RAG-1 and RAG-2 in recombination
• Identify the role of the Artemis enzyme in recombination
• Understand how messenger ribonucleic acid (mRNA) slicing attaches C regions to VJ or VDJ
• Relate junctional diversity to the antigen-combining pocket
• Identify the definition of allelic exclusion
• Explain the role of affinity maturation in immune response
Key Terms
Affinity maturation
Allelic exclusion
Artemis
Combinatorial diversity
Deoxynucleotidyl transferase
Junctional diversity
Ligase
Messenger ribonucleic acid (mRNA) splicing
Nonhomologous end joining
One turn–two turn rule
P-nucleotides and N-nucleotides
RAG enzymes
Recognition signal sequences
Somatic mutation
Signal joints
Introduction
Three theories have been put forth to explain antibody diversity, which allows B cells to generate an antibody repertoire capable of reacting with a wide range of antigens: (1) The germ-line theory postulates that separate genes exist for each antibody molecule and that the antibody repertoire is largely inherited. (2) The deoxyribonucleic acid (DNA) rearrangement theory proposes that a limited number of genes undergo genetic rearrangements to create antibody populations. (3) Finally, the somatic mutation theory proposes that a limited number of inherited genes undergo mutations to general antibody repertoires. In vivo and in vitro studies have demonstrated that both the DNA rearrangement theory and the somatic mutation theory provide the most plausible explanations for antibody diversity.
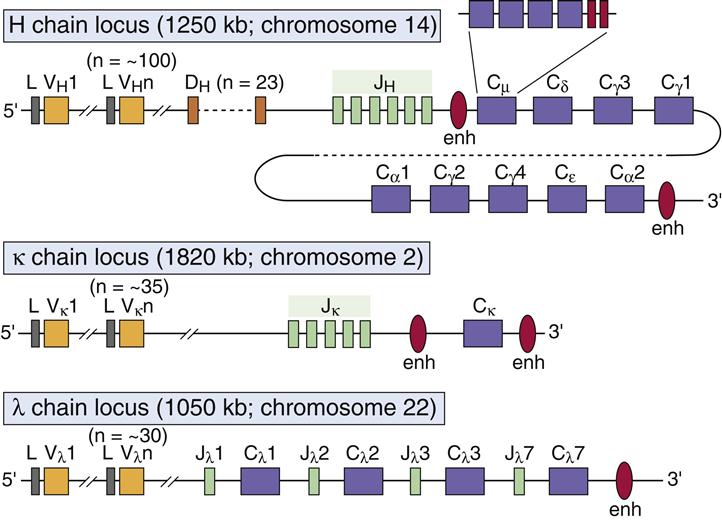
Antibodies are encoded by different germ-line genetic loci. Variable (V) region, joining (J) region, and constant (C) region gene products are assembled into a functional antibody. Variable portion genes (V) code for amino acids that constitute the framework regions of the variable region, and three hypervariable complementarity-determining regions (CDR1, CDR2, and CDR3). The hypervariable regions form the three-dimensional antigen-binding pocket. Antibody specificity is determined by the specific amino acid sequences in CDR3. The joining (J) segment is, in reality, part of the V region and provides some of the framework for the antigen-binding pocket. Only heavy chains have an additional diversity (D) gene.
Antibody diversity is generated from the large number of V, J, D, and C genes available for recombination. Light-chain loci have 30 to 35 genes encoding for the variable (VL) regions (Table 10-1). Five to seven genes code for JL segments in kappa (κ) or lambda (λ) light chains, respectively. Lambda and kappa light chains have one highly conserved constant region.
Table 10-1
Number of Genes Coding for V, D, J, and C Regions in Light and Heavy Chains
| Kappa Light Chains | Lambda Chains | Heavy Chains | |
| Variable segments | 30 | 35 | 100 |
| Diversity segment | 0 | 0 | 23 |
| Joining segment | 5 | 7 | 5 |
| Constant region | 1 | 1 | 5∗ |
| Potential different antibodies | 150 | 245 | 115,000 |
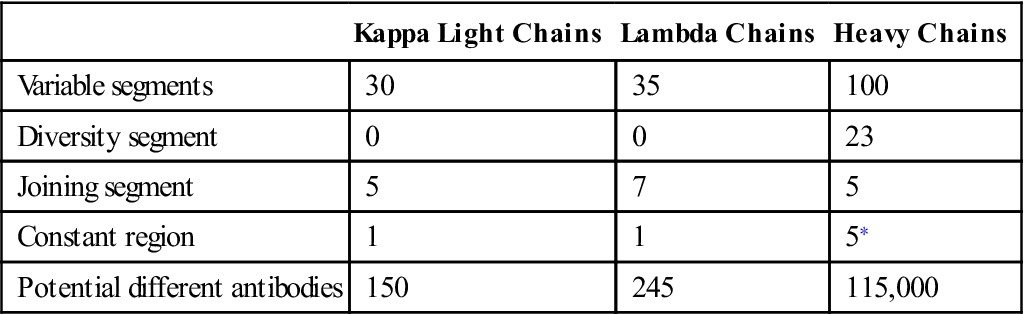
∗Constant region for immunoglobulin G (IgG), IgM, IgA, IgD, and IgE.
Heavy chains are larger than the light chains. A hundred genes code for heavy-chain variable (VH) regions. Diversity (D) genes (N=23) are inserted between V and J genes. J genes (N=5) are linked to the constant region (N=5). The constant region may be from one of the five antibody isotypes (mu [μ], gamma [γ], alpha [α], epsilon [ε], or delta [δ]). An assembled heavy chain consists of VJDC gene products (Figure 10-1).
Combinatorial Diversity
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree