Family of RBPs
C. elegans
D. melanogaster
X. laevis
M. musculus
H. sapiens
PUF
Fbf-1
Pum
Pum1
PUM1
PUM1
Fbf-2
Pum2
PUM2
PUM2
Puf-8
Nanos
Nos-3
Nos
Nanos1
Nanos2
NANOS2
NANOS2
Nanos3
NANOS3
NANOS3
CPEB
Fog-1
Orb
Cpeb1
CPEB1
CPEB1
Cpb-1
Orb2
Cpeb3
CPEB3
CPEB3
Cpb-3
Cpeb4
CPEB4
CPEB4
DAZ
Daz-1
Bol
BOLL
BOLL
Dazl
DAZL
DAZ1 DAZLA
DAZAP
Hrb27C
Dazap1
DAZAP1
DAZAP1
Dazap2
DAZAP2
DAZAP2
2.2 Translational Control of the Mitosis to Meiosis Transition in Drosophila
At the tip of the ovariole in the Drosophila germarium, two germ line stem cells (GSC) divide asymmetrically into a daughter cell which is committed to differentiate and one that replenishes the pool of stems cells (Lin and Spradling 1993; Xie and Spradling 2000). The committed cell, termed cystoblast (CS), undergoes four round of mitosis with incomplete cytokinesis as these cells are connected by cytoplasmic bridges, the ring canals. Two cells of the 16 cell cyst are connected by four cytoplasmic bridges whereas the other cells are connected by three or fewer bridges. One of the cells connected with four bridges will become an oocyte and the other cells will differentiate into nurse cells. Once the oocyte is designated, it receives material from the nurse cells via these ring canals.
Like in nematode gametogenesis, the GSC/CS and mitotic/meiotic transition in Drosophila is regulated by mechanism involving predominantly translational control . Like in worms, Pumilio (Pum) and Nanos (Nos) play a central function in repression of transcripts required for differentiation of the gametes (Miller and Olivas 2011).
The bag-of-marbles (Bam) and benign gonial cell neoplasm (Bgcn) proteins function in both males and females to promote gamete differentiation. Bam is expressed at a critical time when the cystoblast replicates and becomes committed to meiosis entry (Chen et al. 2011b; Slaidina and Lehmann 2014). Bam and Bgcn form a complex that binds and antagonizes both Pumilio and eIF4A proteins. An additional protein included in the complex required for inhibition of GSC factors is Mel-P26, a member of the tripartite motif containing TRIM family of proteins (Li et al. 2013). They also regulate the mRNA coding for another RBP, Nanos , via its 3′UTR (Li et al. 2009; Shen et al. 2009; Kim et al. 2010). Therefore, Bam and Bgcn are part of a network of RBPs that control differentiation of GSCs. A mammalian homolog of Bam was identified and termed GM114 (Tang et al. 2008). However, disruption of this locus has no effect on spermatogenesis (Tang et al. 2008). Since GM114 is remotedly related to Bam, it is still possible that another Bam-ortholog is present in the mammalian genome and involved in translational control during gametogenesis.
2.3 Translational Control of Egg and Embryo Polarity in Drosophila
A fascinating concept based on observation in model organism is that patterning during development is generated by mRNA localization and restricted translation in distinct subcellular domains of a cell. The Drosophila oocyte and embryo is one of the best understood experimental models in terms of development of polarity generated by mRNA localization/translation. In Drosophila the specification of the anterior fate of the embryo is dependent on the localization of maternal mRNA bicoid (bcd) whereas the development of the posterior end depends on the proper localization of osk and nanos (Kugler and Lasko 2009). The localization and translation of these mRNAs generate the anterior/posterior axis. Here we will highlight some very basic concept regarding these mechanisms whereas exhaustive reviews of this process are available (Kugler and Lasko 2009; Ghosh et al. 2012).
Oskar mRNA localization coupled to its translational control is essential for normal development. Osk mRNA is produced by nurse cells and transferred to the oocyte by a complex of proteins including components of the exon junction complex, Staub, and the eIF4E interacting protein Cup and Bruno. Association of the complex with dynein and with microtubules is necessary for the transport. Once in the oocyte, osk localizes in a discrete cytoplasmic structure called the Balbiani body. The site of Oskar protein synthesis determines where germ plasm is assembled. Repression of Osk mRNA translation during this journey is dependent on the eIF4E interacting protein Cup in complex with the RBP Bruno. A Cup ortholog is expressed in mouse oocytes (Clast4) but its function in this species is unknown (Villaescusa et al. 2006). Recently, Clast4 was identified in a screen for genes important for oocyte meiotic reentry although its function was not explored further (Pfender et al, 2015).
Gurken protein is a member of the transforming growth factor-a (TGF-A) family, which functions as ligand for the Drosophila EGF receptor (DER or torpedo). Local mRNA accumulation, local secretion of Gurken and localized occupancy of the EGFR on follicular cells is essential for establishing the antero/posterior axis of the oocyte and the embryo. EGF-like growth factors also play a critical role in oocyte maturation and ovulation in mammals (Conti et al. 2006).
There are two CPEB -like proteins expressed in Drosophila termed Orb (ool8 RNA binding) and Orb2; both of them are required for germ cell development. Orb mutant flies are sterile (Lantz et al. 1994). Null mutant for orb prevents the last replication, and failure to produce oocytes or nurse cells. Later on during differentiation, the Orb protein is localized in the differentiating oocyte indicating an important function in this cell.
Orb undergoes well defined patterns of localization during oocyte development accumulating first in the anterior pole and becoming gradually enriched in the posterior pole of the oocyte. During early embryogenesis, this cytoplasm region will be inherited by primordial germ cells (PGCs). This localization of Orb is necessary for the localization and translation of the osk and Gurken mRNAs. The 3′UTR of osk mRNA plays a critical role in both localization and translation. While osk mRNA is localized to the posterior pole, Gurken mRNA is first deposited to the posterior region of the oocytes and then is translocated together with the nucleus to the anterior dorsal region (Cheung et al. 2011). Orb mutations interfere with both localization processes. Gurken acts both early and late in the oocyte assembly process (Cheung et al. 2011). Consequently, since Orb is involved in both phases of Gurken mRNA localization , Orb also acts both early and late in oocyte assembly (Christerson 1994). From the mechanistic standpoint, it has been shown that Orb and poly-adenosine polymerase (Pap) control the polyadenylation and translation of oskar mRNA confirming their function as translational activators.
2.4 Translational Control of the Meiotic Cell Cycle in Danio Rerio Oocytes
Oocyte maturation in zebrafish is triggered by MIH (17a,20b-Dihydroxy-4-pregnen-3-one) and governed by activation of pre-MPF, which consists of cyclin B and inactive Cdk1 (Brownlie et al. 2003). As with Drosophila, some mRNAs are deposited in zebrafish oocytes in a polarized fashion. Cyclin B mRNA is concentrated in RNA granules along the cytoplasm at the animal pole through an interaction with the RNA binding protein Pumilio1. Formation of RNA granules depends on actin filaments since cytochalasin treatment leads to cyclin B mRNA release from the granules. After stimulation by MIH, the cyclin B mRNA disperses into the cytoplasm, facilitating its translational activation (Kotani et al. 2013). Cyclin B mRNA localization within the oocyte is dependent on sequence elements within the open reading frame (ORF) as well as the 3′UTR (Yasuda et al. 2010).
As in other species, zebrafish cyclin B mRNA is regulated by RBPs. Recently Pumilo and Igf2bp3 (insulin -like growth factor 2 mRNA-binding protein 3) were reported to repress translation by binding the 3′UTR of cyclin B (Takahashi et al. 2014; Kotani et al. 2013). Compared to other model organisms CPEB dependent translation of cyclin B mRNA is considerably less efficient, probably due to different positioning and sequence of the CPEs (Zhang and Sheets 2009), indicating that the optimized level of cyclin B for the oocyte maturation is species specific. Zebrafish oocytes express two classes of CPEBs, the oocyte specific Zorba together with the embryonic type ElrA (O’Connell et al. 2014). Also Zorba mRNA localizes at the animal pole at stage II of oogenesis (GV stage). This localization seems to be independent of microtubules and microfilaments (Bally-Cuif et al. 1998).
2.5 Translational Control of the Meiotic Cell Cycle in Xenopus laevis Oocytes
Frog oocytes have been a model used for extensive studies on the role of translational control of germ cells. Cpeb was first described in frog oocytes and much of what is known about the function of this protein comes from studies in Xenopus. In frogs like in most species studied, maternal mRNAs accumulate during oocyte growth but they are not translated into proteins (Clarke 2012). The polyadenylation state of these mRNAs is thought to be central to the repressed state and for translational activation (Richter 2007; Charlesworth et al. 2013). In frog oocytes, a large macromolecular complex is formed involving RBPs (Cpeb1, Cpsf, ePab), an adenylase (Gld2), a deadenylase (Parn), and the scaffold/adaptors Symplekin and Maskin , as well as components of the cap complex (eIF4E, eIF4E-T). This complex is thought to prevent translation by maintaining a short poly(A) tail (Fernandez-Miranda and Mendez 2012; Ivshina et al. 2014). Other complexes have been described as important for repression, including Pumilio proteins (Kim and Richter 2007) or Cup/Orb in Drosophila (Wong and Schedl 2011). Further function of these complexes is discussed below in the context of mammalian oocyte.
3 RBPs and Translational Control in Mammalian Primordial Germ Cells (PGCs)
In the mouse fetal gonad, male PGCs exit mitotic cycle and enter a period of quiescence, whereas female PGCs become committed to enter meiosis. The signal for this decision is extrinsic and derives from somatic cells. It is commonly accepted that retinoic acid (RA) induces female germ cells to enter meiosis and that this signal is suppressed in the male through rapid metabolism of RA (Bowles et al. 2006). A key regulator of meiotic cell cycle entry is stimulated by retinoic acid 8 (Stra8) with probable function of transcription regulator. However, RBPs play a key role in the control of Stra8 expression. Nanos 2 and 3 proteins are essential for PGC development in a dymorphic fashion with Nanos 2 expressed only in male germ cells. Nanos 2 function is to promote the male fate but repress the female fate in germ cells (Saga 2010). Nanos acts as a translational repressor that prevents meiotic entry by suppressing translation of several mRNAs including Stra8 itself.
The RBP DAZL (deleted in Azoospermia-like) is also considered an important component required for entry into meiosis. It has been proposed that DAZL has a permissive function as a “licensing” factor, priming germ cells to respond to molecular cues and engage germ cells in sex differentiation (Gill et al. 2011) but the molecular mechanisms involved are unclear. Traditionally DAZL is viewed as an activator of translation but it has been recently proposed that it may also function as repressor. Indeed DAZL interacts with Pumilio proteins which are usually repressor of translation (Urano et al. 2005). CPEB1 also plays a role because CPEB1 knockout prevents PGC entry into meiosis and SYCP3 expression (Tay and Richter 2001). CPEB is necessary for DAZL protein accumulation and it is likely that CPEB1 and DAZL function sequentially and synergistically during female PGC commitment to enter meiosis.
4 Translation Control during Adult Mammalian Spermatogenesis
In the adult mammalian male gonad, the production of gametes is divided into three stages: a proliferation stage, where spermatogonia undergo five rounds of mitotic division (Oatley and Brinster 2008; Hermo et al. 2009), a maturation phase, where diploid spermatocytes undergo two meiotic divisions to yield haploid spermatids (Zickler and Kleckner 1999) and spermiogenesis where haploid spermatids differentiate into spermatozoa (Braun 2001). The progression through each of these events relies on a tightly controlled network of specific genes regulated at the transcription and post-transcriptional level.
The temporal regulation of the protamine 1 (Prm1) mRNA translation during spermiogenesis is a classic example of translational regulation and uncoupling of transcription and translation. Transcription of this mRNA initiates during the round spermatid stage (Braun et al. 1989) but the mRNA is not translated immediately. The translation of the Prm1 mRNA is dependent on a highly conserved sequence, the translational repression being mediated by a 17-nucleotide translational control element (TCE) located in the 3′UTR of this mRNA. Mutation of this TCE cis-acting element causes premature synthesis of protamine protein and sterility. The Prm1 mRNA is stored as a cytoplasmic ribonucleoprotein (mRNP ) particle in spermatids that are also required for maintaining this repressed state. Translational activation of stored Prm1 mRNA in elongated spermatids requires the trans-activation responsive RNA-binding protein 2 (TARBP2). Mice mutant for Tarbp2 are defective in proper translational activation of the Prm1 and Prm2 mRNAs and are sterile (Zhong et al., 1999). Tarbp2 is expressed at high levels in post-meiotic spermatids. The cold shock domain Mouse Y-box protein 2 (MSY2) is one of the proteins responsible for repression of Prm1. This is believed to prime transcripts (with a Y-box region within their promoter) to cytoplasmic repression and RNA stability (Yang et al. 2005). In MSY2 knockout models, no spermatozoa are found in the seminiferous tubules, in part due to reduced chromatin condensation (Yang et al. 2005)
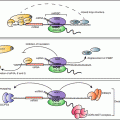
Additional RBPs have been described both in the nuclear and cytoplasmic components of cells, some of which shuttle between the nucleus and cytoplasm. Once in the cytoplasm, transcripts are encapsulated in RNPs stabilized by RBP protein–protein and protein–RNA interactions. The effect on mRNA processing by RBPs is further augmented by post-transcriptional modifications of the RBPs (methylation, ubiquitination, phosphorylation, acetylation) that remodel the structure of RNPs and alter their activity (Bettegowda and Wilkinson 2010; Idler and Yan 2012). For example, knockdown of the KH domain-containing protein SAM68 results in infertility associated with germ cell apoptosis and aberrant elongating spermatids. This protein binds polyadenylated transcripts with the AUUAAA motif and upon phosphorylation translocates from the nucleus to the cytoplasm promoting function of signal transduction, alternative splicing and translation upregulation (Lin et al. 1997; Najib et al. 2005; Paronetto et al. 2009; Paronetto and Sette 2010). Here we will focus on the properties and function of four classes of RBPs that are critical for spermatogenesis progression (Fig. 7.1).
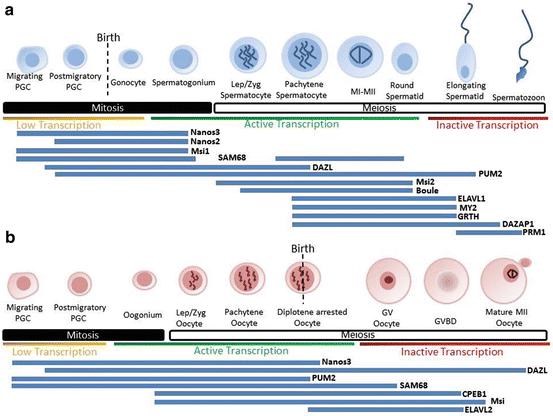

Fig. 7.1
Expression profile of RBPs throughout mouse spermatogenesis (a) and oogenesis (b). Progression through gametogenesis is highly regulated at the translational level, both in the mitotic and meiotic phases. Developmental progression is regulated by complex network of RBPs which control, translation activation, repression and RNA stability. The following references contain details on the expression of genes expressed during spermatogenesis and ovogenesis: Pum2 (Xu et al. 2007), Nanos 2 (Barrios et al. 2010), Nanos 3 (Lolicato et al. 2008), Dazl (Ruggiu et al. 1997; Saunders et al. 2003; Chen et al. 2011a), Boule (Xu et al. 2001). Sam68 (Paronetto et al. 2009), Elavl2 (Chalupnikova et al. 2014), CPEB1 (Tay and Richter 2001; Chen et al. 2011a), Musashi (Gunter and McLaughlin 2011)
4.1 Deleted in Azoospermia Family of RBP
The germ cell specific Deleted in Azoospermia (DAZ) family of proteins is composed of three members; the primate and catarrhine lineage specific protein DAZ, located within the long arm of the Y chromosome (Reijo et al. 1995); the autosomal and family ancestor Boule, with homologs described from flies and worms to mice and humans (Karashima et al. 2000); and the vertebrate specific DAZ-like (DAZL) (Bielawski and Yang 2001). This family of proteins owes its name to the phenotype of patients were it was first identified. In a genetic screen of azoospermic men, 14 % of cases had deletions within the area containing the DAZ coding region (Reijo et al. 1995). The other members of the DAZ family have also been associated with male infertility as reported in several SNPs studies (Chen et al. 2010).
All members of the DAZ family are required during spermatogenesis. DAZ and Boule are necessary for spermatid development, with Boule-null mice showing a developmental arrest at step 6 of spermatids (VanGompel and Xu 2010). DAZL phenotype can be detected much earlier compared to its orthologues, with a spermatogenic arrest at prophase of meiosis I (Ruggiu et al. 1997; Saunders et al. 2003). This phenotype becomes more penetrant in a pure BL6/C57 background, with apoptosis observed at embryonic day 15.5 of mouse development (Lin and Page 2005). Genetic studies have associated this phenotype with an impairment of a canonical PGC feature, sex differentiation.
The nuclear and cytoplasmic localization of DAZL (Xu et al. 2001), the ability to bind mRNA (Tsui et al. 2000b; Venables et al. 2001), the association with polyribosomes (Tsui et al. 2000b) and the interaction with several known RBPs (Brook et al. 2009) suggest a role in mRNA processing. Several molecular functions have been associated with DAZL, ranging from mRNA repression (Urano et al. 2005; Padmanabhan and Richter 2006) mRNA transport (Lee et al. 2006; Kim et al. 2012), miRNA-mediated repression protection (Takeda et al. 2009), but to date the best characterized role for DAZL is in translation activation (Collier et al. 2005; Reynolds et al. 2005, 2007; Chen et al. 2011a). In vitro studies have demonstrated that DAZ family members all share a common function as translation activators. Tethering assays, where DAZL-RNA binding activity is circumvented, showed that this family upregulates translation of artificially bound luciferase reporters (Collier et al. 2005). In vivo experiments have since demonstrated translation of endogenous transcripts (Reynolds et al. 2005, 2007; Chen et al. 2011a). Insight into the mechanism of DAZ-family translation activation demonstrates that these proteins act at the step of translation initiation. A direct interaction with PABP1 (but none of the other canonical translation initiation factors) was shown to be necessary to promote translation in a reconstitution system. The observation that DAZL was able to stimulate translation of non-adenylated reporter mRNAs indicated that this can work distinctly from the canonical CPEB translation activator (Pique et al. 2008) by promoting PABP recruitment in a poly(A) independent manner (Collier et al. 2005).
DAZL interacts with transcripts with a U-rich 3′UTR (Tsui et al. 2000b). A variety of assays identified the trinucleotide GUU has the minimal binding motif of DAZL (Tsui et al. 2000b; Ruggiu and Cooke 2000; Venables et al. 2001). The crystal structure of DAZL RNA recognition motif (RRM) demonstrated that this presents the highest affinity towards GUU[U/C] sequences (Jenkins et al. 2011). The relatively short sequence of this element greatly challenges the identification of DAZL targets. In vitro evidence for the requirement of multiple DAZL binding sites for a maximal translation activation can in the future help to overcome this challenge (Collier et al. 2005; Chen et al. 2011a). Several studies have identified DAZL targets (Brook et al. 2009). In spermatogenesis, pull down experiments have demonstrated an interaction with messages involved in sperm development (Tpx1, Grsf-1, Trf-2, TssK proteins, etc.) (Jiao et al. 2002; Zeng et al. 2008). Nonetheless, to date the only two bona fide spermatogenic targets for DAZL are Sycp3 and Mvh (Reynolds et al. 2005, 2007).
4.2 Pumilio Family of RBPs
Whereas the role of Puf proteins in model organisms have been extensively investigated, comparable little information is available on the role of Pumilio proteins, Pum1 and Pum2, in spermatogenesis and oogenesis in mammals. A gene trap disruption of Pum2 was reported to produce a decrease in testis weight and some disruption in spermatogenesis but the exact steps affected were not investigated (Xu et al. 2007). Pum1 knockout mice have decreased fertility and decreased sperm count and 25–30 % reduction in testis weight. It has been proposed that this is due to deregulation of the apoptotic pathway in the Pum1−/− mice during spermatogenesis (Chen et al. 2012). Pum1-dependent repression of Map3k1 is required to maintain P53 inactive. Since Pum1 targets a large number of mRNAs in cell lines and in the testis it is most likely that Pum1 functions go well beyond P53 regulation. In model organism, Pumilios play a critical role in maintaining the germ cell fate and prevent germ cell differentiation. It may be possible that accelerated differentiation of spermatogenic cells in the Pum1 knockout model leads to unbalanced ratio/germ cell supporting cells in the testis, a condition known to trigger apoptosis and spermatogenic failure.
4.3 Deleted in Azoospermia Associated Protein 1 (DAZAP1)
DAZAP1 is a member of the heterogeneous nuclear ribonuclear protein family (hnRNP). This protein was initially identified in a study looking at DAZL interacting proteins (Tsui et al. 2000a). But despite this interaction, both proteins differ in their protein expression pattern and were described to be associated with different subsets of target mRNAs, indicating that both proteins can have different roles (Kurihara et al. 2004).
DAZAP1 is ubiquitously expressed but highly enriched in testes (Dai et al. 2001). Whereas it is predominantly found in the nucleus of somatic cells (Lin and Yen 2006), in testes, DAZAP1 shows a dynamic distribution. In germ cells DAZAP1 is found in the nucleus from mid-pachytene spermatocytes to round spermatids, and relocates to the cytoplasm in elongating spermatids (Vera et al. 2002).
As with other hnRNPs, DAZAP1 binds newly synthesized transcripts and, through processes of splicing, export and translation, controls expression of specific messages (Martinez-Contreras et al. 2007). Recent evidence further elucidated the mechanism employed by DAZAP1 in splicing control based on extracellular cues. Through interaction with hnRNPs governed by 3′UTR cis-elements of target transcripts, DAZAP1 promotes splicing of weak exons. Phosphorylation of the C-terminal proline rich domain of DAZAP1 by MEK/Erk pathway alters the ability of DAZAP1 to maintain protein interactions with splicing factors and induces cytoplasmic translocation of DAZAP1 (Choudhury et al. 2014).
The molecular mechanisms of translational modulation of DAZAP1 are still not fully understood. It binds RNA at a consensus sequence AAAUAG and GU[1–3]AG (Yang et al. 2009), and the function is dependent on the protein partners to which it binds. Protein–protein interactions have been described with DAZL, KH-type splicing regulatory protein (KHSRP), hnRNPs, DEAD box polypeptide 20 (DDX20), let 7 and many other RBPs (Yang et al. 2009). The function as a translation activator has been studied in vitro models where DAZAP1 proteins were tethered to luciferase reporters. These show that both mouse and human DAZAP, as observed for X. laevis, stimulates translation in a cap-independent manner and it shows preferential translation towards nonadenylated messages (Smith et al. 2011).
Interestingly, DAZAP1 is itself a target for translation control. Although initially present at mid-pachytene spermatocytes, in situ analysis of DAZAP1 detects high levels of this transcript in spermatogonia and early spermatocytes (Vera et al. 2002). Through alternative usage of cleavage and polyadenylation sites, translation of each isoform is regulated by different mechanisms. At day 12 post-partum association with polyribosomes shows that both isoforms are actively translated. Upon reaching puberty the isoform with the longest 3′UTR relocates to the translational inactive mRNP fraction with a concomitant shortening of the poly(A) tail. The smaller isoform is also recruited to the mRNP fraction but this shows no changes in poly(A) length. The germ cell specific DAZL was shown to preferentially bind in vivo to the shorter isoform of DAZAP1, and in vitro assays also described a preferential DAZL promoted translation of a reporter with the 3′UTR of the shorter isoform (Yang and Yen 2013).
4.4 Gonadotropin-Regulated Testicular RNA Helicase (GRTH)
Ddx25 gained its common name, GRTH, from the fact that translation of this protein is dependent on hormonal cues. Three consensus Kozak sequence s can be identified in frame with the coding region of GRTH. Whilst the second AUG is used by Leydig cells, germ cells preferentially use the first and third start codons. Although, treatment of rats with hCG, resulted in a shift towards the second AUG by round spermatids, but not spermatocytes (Sheng et al. 2003). This translational control is believed to result from induced paracrine factors that regulate the internal ribosomal entry site mechanisms.
GRTH promotes survival of spermatocytes by regulating apoptotic pathways. In Grth-null models, spermatogenesis arrests at step 8 spermatids due to increased apoptosis in spermatocytes at stage XII (Tsai-Morris et al. 2008). Despite not totally understood how GRTH regulates translation, an initial mechanism was directly linked to the function of RNA helicases that control RNA unwinding and nuclear transport (Tsai-Morris et al. 2010). An association with chromatoid bodies and polyribosomes further supported a role of this protein in translation (Tang et al. 1999; Sheng et al. 2006; Tsai-Morris et al. 2004b). Animal models provided the direct link to translation control by providing evidence that decrease levels of GRTH in germ cells resulted in decreased levels of specific proteins (TNPs and tACE) with no differences observed in mRNA levels (Tsai-Morris et al. 2004a). GRTH is present in the nucleus and cytoplasm, and is coupled with nuclear export, dependent on its phosphorylation status. Depletion of GRTH resulting in smaller chromatoid bodies and a deficit in the cytoplasmic ratios of proteins associated with nuclear histone-protamine (Tsai-Morris et al. 2004b; Sheng et al. 2006).
In humans, a mutation on GRTH is associated with non-obstructive azoospermia. This mutation results in a nonphosphorilated form of GRTH, and was proposed to impair RNA-binding and protein–protein interactions due to the location of the substituted residue in a hydrophobic pocket (Tsai-Morris et al. 2007).
5 Regulation of Translation in Mammalian Oocytes
The post-natal ovary is endowed with a pool of quiescent primordial follicles. Upon primordial follicle activation, the oocyte enters a growth phase that is characterized by intense transcriptional activity (Pan et al. 2005). However not all the transcribed mRNAs are immediately translated; a fraction is stored in a translationally-repressed form. As the oocyte approaches its fully grown size, the transcriptional activity ceases (Bouniol-Baly et al. 1999; Liu and Aoki 2002) and the final stages of differentiation and maturation, as well as fertilization and early embryo development occur in absence of transcription. Thus, the meiotic cell cycle progression and genome reprogramming rely on unmasking and translation of stored maternal mRNAs. This dependence continues until the embryo genome becomes activated, an event that occurs with a timing that is species specific (2-cell stage for major embryonic genome activation (EGA) in mice Latham 1999). As mentioned, this unique control of gene expression at the oocyte-to-embryo transition likely confers developmental plasticity by regulating transcript recruitment and translation in the cytoplasm, while the chromatin is free to undergo the reprogramming required for establishing totipotency and supporting embryogenesis.
Here we will present the most relevant mechanisms involved in the regulation of maternal mRNA translation, organizing the discussion into mechanisms controlling the activation, degradation and repression during oocyte development.
5.1 Mechanisms Controlling Repression of Translation and mRNA Degradation
Our current understanding of translational regulation during oocyte growth and maturation is mainly based on findings in the X. laevis oocyte model. In the frog system, it has been proposed that a combinatorial code of cis-acting elements in the 3′UTR of the messages regulates protein synthesis according to the temporal requirement of meiosis arrest and progression, as well as early embryo development.
One of the best characterized mechanisms of translation regulation in frog oocyte requires the cis-acting element CPE and the cognate binding proteins CPEB . By controlling the length of the cytoplasmic poly(A) tail, Cpeb1 represses translation in immature oocytes and, when phosphorylated upon cell cycle reentry, activates translation (McGrew et al. 1989). A mammalian isoform of CPEB, with 80 % identity with the frog Cpeb, was first described in mouse oocytes as a protein bound to the c-Mos mRNA (Gebauer and Richter 1996). Early studies of the tissue-type plasminogen activator (tPA) mRNA provided initial evidence that maternal mRNA repression and translation in mammals are regulated similarly to frogs (Huarte et al. 1992; Stutz et al. 1997). For instance, tPA is stored in a silenced form with a short poly(A) tail (30–40 nt) in GV stage oocytes. Upon meiotic re-entry, the poly(A) tail is elongated (to about 200 nt) and tPA becomes translated (Huarte et al. 1992). The role of a CPE cis-acting element was soon recognized as an important player in this regulation. By inserting a CPE or deleting an existing one, the translation is respectively repressed or activated, together with shortening or lengthening of the poly(A) tail (Huarte et al. 1992; Gebauer and Richter 1996).
How CPEB functions in repressing poly(A) elongation and translation of maternal transcripts in mammals is still not fully understood. At least three different models have been proposed in frog oocytes, depending on the composition of the repressor complex, as reviewed in (Villalba et al. 2011). One possibility is that Cpeb simultaneously recruits the poly(A)-ribonuclease Parn and the poly(A)-polymerase Gld-2. The competition between the two enzymatic activities maintains a short poly(A) tail, hence promoting repression (Kim and Richter 2006). Alternatively a repressor protein, identified as Maskin , may bind to Cpeb and eIF4E, blocking the formation of the cap-binding complex on the mRNA, resulting in translational inhibition (Stebbins-Boaz et al. 1999). Similarly, a third model predicts that Cpeb bind to Clast 4 (also known as 4E-transporter (4E-T)), which in turn recruits isoforms of eIF4E, like eIF4E1b, that have low affinity for the cap structure (Minshall et al. 2007).
Rather than being mutually exclusive, these models may be integrated in the control of translational repression of different mRNA species or during different steps of oogenesis. Moreover extra layers of complexity are achieved through multiple regulatory cis and trans-acting factors. For instance additional regulation can be mediated by Pumilio binding elements (PBEs) that are frequently present in mRNAs that possess CPEs (Ota et al. 2011). As previously discussed in model organisms, Pumilio proteins (PUM1, PUM2) are mainly repressor proteins, but they can also stabilize CPEB binding and promote poly(A) tail elongation in some context (Padmanabhan and Richter 2006; Pique et al. 2008). However their role in mammalian oogenesis has not been elucidated yet. Interestingly, it has been reported that Pum1 protein in zebrafish oocytes is required to localize cyclin B1 to ribonuclear particles and this co-localization was confirmed in mouse oocytes (Kotani et al. 2013).
Another cis-acting element known to repress translation in immature oocytes is the translational control sequence (TCS). The corresponding trans-acting factor has been recently discovered in X. leavis and it is represented by zygote arrest 1 (Zar1) and Zar2. Zar1 and Zar2 bind to Mos and Wee1 mRNA 3′UTRs via a zinc finger. The TCS-mediated repression of translation is exerted during oocyte growth and initial phases of meiotic resumption until the metaphase I (MI) stage (Charlesworth et al. 2012; Yamamoto et al. 2013). It is not clear if the translational activation of Zar2 target mRNAs occurs following partial degradation of Zar2 and release of the repressive state, or if the control requires more than one trans-acting factor, as proposed for CPE. Based on the observation that mice embryos that are null for Zar1 arrest at the 1-cell stage due to failure in EGA, mammalian ZAR family proteins were initially proposed to be transcriptional regulators (Wu et al. 2003). However, a role for ZAR proteins in translational control in mammalian oocytes cannot be excluded. In this view, protein product(s) of ZAR-target mRNAs would be responsible of EGA, and the absence of ZAR would prevent their timely translation with subsequent failure in embryonic transcription and development.
In X. laevis oocytes CPE-induced translation is responsible for activating synthesis of RBPs, like the zinc finger protein C3H-4, that are required in the later stages of maturation for deadenylation of transcripts carrying A-U rich elements (AREs) (Belloc and Mendez 2008). In mammals, the embryonic lethal abnormal vision like 2 (ELAVL2) protein regulates ARE -mediated mRNA repression during oocyte growth (Chalupnikova et al. 2014). ELAVL2 decreased in abundance as the oocyte reaches a fully grown, meiotically competent stage of development, which is characterized by the transition from the non-surrounded nucleolus (NSN) chromatin configuration into the transcriptionally quiescent surrounded nucleolus (SN) configuration. ELAVL2 is absent in MII oocytes and zygotes. Mammalian proteins that function analogous to the frog C3H-4 protein to mediate ARE dependent deadenylation and repression during oocyte maturation have not been identified.
A regulatory feedback involving CPE-mediated translation to recruit decapping mRNA1 and 2 (Dcp1a and Dpc2) has been recently described in mice (Ma et al. 2013). Decapping , i.e. the removal of the 5′ 7-methyl-guanosine cap, exposes mRNAs to exonucleases and represents a critical step in the 5′→3′ transcript degradation. Both the regulatory and catalytic subunits, DCP1A and DCP2, are synthesized between MI and MII and the inhibition of their accumulation interferes with the proper EGA execution. It is not clear at this point whether and how the DCP1A/DCP2-mediated degradation could be selective. One of the possibilities is that their involvement is the downstream event in a multi-step process that first requires mRNAs to be destabilized and tagged for degradation by sequence specific regulatory factors.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree