Fig. 22.1
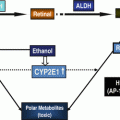
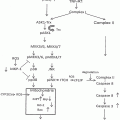
Major enzymatic pathways involved in ethanol and glutathione metabolism. Ethanol (EtOH) is subject to metabolism by catalase (CAT), cytochrome P450 isoform 2E1 (CYP2E1), and alcohol dehydrogenase (ADH). Acetaldehyde is metabolized by aldehyde dehydrogenase (ALDH) isoforms 1A1, 1B1, and 2. In the glutathione (GSH) pathway, glutamate cysteine ligase (GCL), which includes two subunits the catalytic subunit (GCLC) and the modifier subunit (GCLM), catalyzes the synthesis of γ-glutamylcysteine (γ-GC). γ-GC is then coupled to glycine by glutathione synthetase (GSS) to form GSH. During oxidative processes, reactive oxygen species (ROS) form which can cause lipid peroxidation. ROS can be reduced by GSH, in the process forming GSSG, the oxidized form
Taken together, the association between human polymorphisms of ethanol-metabolizing genes and alcohol-related diseases implicates a significant pathogenic role of ethanol metabolism in alcohol toxicity. Various research groups have developed animal models that harbor genetic ablations of ethanol-metabolizing enzymes. These models can serve as important experimental tools to elucidate the mechanistic roles of specific enzymes or pathways in alcohol-related diseases and therefore have direct relevance to alcohol research.
22.3 Animal Models for Alcohol-Induced Cancer
Low to moderate consumption of alcohol has tissue-protective properties [33]. However, heavy alcohol consumption increases the risk of several diseases, including cancer. A comprehensive review of epidemiological data demonstrated a significant increase in cancer risk for several epithelial-derived tumors associated with ethanol consumption [3]. Studies using experimental animals, however, support the notion that ethanol acts as a cocarcinogen or tumor promoter rather than being a carcinogen itself [5]. The mechanisms by which alcohol promotes tumorigenesis remain unclear due primarily to a lack of good animal models that can recapitulate the increased risk of alcohol in carcinogenesis. Animal models analogous to inflammation-promoted cancers, such as the azoxymethane (AOM)/dextran sulfate sodium (DSS) model, are needed [34]. In this model, the carcinogen AOM by itself is administered at a dose that causes no dysplasia; however, when administered in combination with DSS (which induces inflammation), a synergistic increase in the number of tumors is observed. The creation of a similar model for alcohol will rely on a better understanding of the interactions between the genetic mutations (or carcinogen) with the duration, route, and concentration of ethanol for the epithelial tumor that is to be modeled. Moreover, diets that better mimic heavy alcohol consumption in humans are required. The interaction of tumor-promoting dietary components, such as alcohol, high-fat, and iron, may lead to more robust and precise models. Lastly, experimental evidence indicates that the metabolism of ethanol leading to the generation of acetaldehyde and free radicals is intimately involved in alcohol-associated carcinogenesis [3]. Therefore, a more comprehensive understanding of the enzymes required for alcohol metabolism in cancer are needed and the genetic animal models discussed in this review could represent unique opportunities to identify their roles in alcohol-induced cancers.
22.4 Glutathione in Alcoholic Tissue Injury
In the development of alcohol-induced tissue injury, it is apparent that numerous pathways in target organs are modulated by ethanol [35, 36]. Oxidative stress appears to play a central role in many of these pathways [37]. Ethanol metabolism, CYP2E1 induction, compromised antioxidant defense, mitochondrial injury, inflammation, hypoxia, and iron overload can all contribute to the alcohol-induced oxidative environment. Accumulation of the reactive molecules (including reactive oxygen species and electrophilic products, such as acetaldehyde and lipid peroxidation products) can be harmful to a biological system due to their propensity to inactivate enzymes and cause DNA damage, loss of protein functions and cell death [38].
Glutathione (GSH) plays an important role as an antioxidant by serving as a cofactor for antioxidant enzymes, such as glutathione peroxidase and glutathione S-transferases, or by directly scavenging free radicals [39]. It is the most abundant nonprotein thiol, attaining a concentration in the high millimolar range in the liver [40]. Because of its abundance, GSH plays a key role in maintaining cellular redox homeostasis and, therefore, enzymes that help generate GSH are critical in protecting cells against oxidative stress. GSH is a tripeptide composed of glutamate, cysteine, and glycine. It is synthesized in most types of cells by two successive enzymatic reactions. The first reaction couples glutamate and cysteine and is catalyzed by glutamate-cysteine ligase (GCL), resulting in the formation of γ-glutamylcysteine (γ-GC) [41] (Fig. 22.1). The second reaction, catalyzed by GSH synthetase, couples γ-GC with glycine. The formation of γ-GC by GCL is considered rate-limiting in GSH biosynthesis, and GCL has been the principal target of drugs designed to inhibit GSH biosynthesis [41] and to generate mice with GSH deficiency [42].
In higher eukaryotes, GCL in its most catalytically efficient form is a heterodimer composed of a catalytic (GCLC) and a modifier (GCLM) subunit, each of which is encoded by separate genes on different chromosomes. GCLC possesses all of the catalytic activity of γ-GC formation; GCLM optimizes the kinetic properties of the holoenzyme, thereby regulating tissue GSH levels [43]. GSH is exclusively synthesized in the cytoplasm [39] and further distributed into mitochondria, endoplasmic reticulum, and nuclei, where it plays a pivotal role in the normal functioning of these subcellular organelles [44]. During detoxication of free radicals, GSH is oxidized to glutathione disulfide (GSSG). Both GSH and GSSG can be transported outside the cell where it is broken down in sequence by γ-glutamyl transferases and dipeptidase, producing free cysteine and glycine for intracellular reutilization [45]. Depletion of hepatic GSH, particularly mitochondrial GSH, occurs as a result of excessive GSH consumption by free radicals and acetaldehyde generated during alcohol metabolism [46]. Given the above considerations, animal models exhibiting a GSH deficiency will serve as important tools to study GSH-regulated redox biology in ethanol metabolism and ethanol-induced tissue damage. As such, they are of direct relevance to alcohol research.
22.5 Mouse Models with Genetic Deficiencies in Ethanol-Metabolizing Enzymes
ADH1 global knockout: The Adh1 −/− mouse line has been generated by Duester [47] (Table 22.1). It should be noted that human ADH1 gene family consists of three genes, viz. ADH1A, ADH1B, and ADH1C, whereas the mouse genome has a single Adh1 gene [11]. Adh1 −/− mice have limited capacity to oxidize ethanol and retinol. Pharmacokinetic studies show a reduction in blood ethanol clearance in these animals [9]. Following parenteral administration of ethanol, these mice displayed an increased sleep time and embryonic resorption was increased threefold [9]. While ADH1 is thought to be responsible for the majority of ethanol metabolism in the liver, new pharmacokinetic evidence suggests a role for other ADH isoforms as well [48]. Therefore, this model may be useful in determining the pathophysiological importance of compensatory ADH isoforms as well as elucidating the kinetics of these enzymes for ethanol.
Table 22.1
Transgenic mouse models
Strain | Genetic background | Phenotype | References |
|---|---|---|---|
Adh1 −/− | C57BL6 | • No gross abnormality | Deltour et al. [9] |
• Reduction in blood ethanol clearance | |||
Cat −/− | C57BL6 | • No gross abnormality | Ho et al. [49] |
• Deficiency in brain mitochondrial respiration | |||
• Has not been used in ethanol toxicity studies | |||
Cyp2e1 −/− | 129/Sv | • No gross abnormality | |
• Decreased sensitivity to acetaminophen hepatotoxicity | |||
• Resistance to ethanol-induced fatty liver and oxidant stress | |||
Cat −/− Cyp2e1 −/− double knockout | C57BL6/129 mixed | • No gross abnormality | Unpublished |
• Has not been used in ethanol toxicity studies | |||
Aldh2 −/− | C57BL6 | • No gross abnormality | |
• High susceptibility to ethanol toxicities by oral administration | |||
• High sensitivity to inhalation toxicities of acetaldehyde | |||
Aldh1a1 −/− | C57BL6 | • Viable and fertile | Fan et al. [74], Ziouzenkova et al. (2007) |
• Decreased susceptibility to diet-induced obesity and insulin resistance | |||
• Cataract development at age of 6-month | |||
• Has not been used in ethanol toxicity studies | |||
Aldh1b1 −/− | C57BL6 | • No gross abnormality | Unpublished |
• Reduction in blood acetaldehyde clearance |
Catalase global knockout: The Cat −/− mouse strain was developed and characterized by Ho and colleagues [49] (Table 22.1). These mice do not express catalase and develop normally, i.e., exhibit no gross abnormalities. However, brain mitochondria of these animals show deficiencies in mitochondrial respiration. To date, this knockout strain has not been subjected to ethanol toxicity studies. Given that earlier studies have shown a significant role of catalase in modulating ethanol sensitivity in the brain [14, 15], the Cat −/− mice would be anticipated to be a valuable animal model for examining ethanol drinking preference as well as alcohol toxicities.
CYP2E1 global knockout: CYP2E1 is an ethanol-inducible enzyme with a role in hepatic ethanol oxidation. By genetically ablating exon 2 of Cyp2e1 gene, Gonzalez and colleagues developed Cyp2e1 –/– mice [50] (Table 22.1). These mice do not express CYP2E1 enzyme and develop normally [50]. They also show lower sensitivity to the deleterious hepatic effects of acetaminophen [50]. As one of the primary xenobiotic/endobiotic-metabolizing p450s, CYP2E1 is a contributor to a variety of cellular toxicities induced by endogenous or exogenous pathogens. Using the Cyp2e1 –/– mouse model, CYP2E1 has been shown to play a pivotal role in mediating hepatotoxicity making this an interesting model for alcohol-related liver toxicity [51, 52] Cyp2e1 –/– and Cyp2e1 knock-in mice have been used to examine the potentiation of ethanol-induced hypoxia. Cyp2e1 –/– mice exhibited the lowest levels of hypoxia and HIF1-α [53]. Similarly, ethanol-induced fatty liver and oxidant stress are blunted in these mice [54]; this study confirmed the important role of CYP2E1 in ethanol-induced liver toxicities. Cyp2e1 −/− mice also display longer ethanol-induced sleep time than do wild-type mice [15], confirming the relevance of the Cyp2e1 −/− mouse line for the study of the CYP2E1 enzyme in ethanol toxicities and alcohol-induced drinking preference.
22.6 Mouse Models with Genetic Deficiencies in Acetaldehyde-Metabolizing Enzymes
ALDH2 global knockout: The Aldh2 −/− strain was originally developed and characterized by Isse and colleagues [55, 56] (Table 22.1). Aldh2 −/− mice do not express ALDH2 protein and have no detectable capacity to oxidize acetaldehyde, propionaldehyde, or methoxyacetaldehyde in liver mitochondrial fractions. Following oral administration of ethanol, Aldh2 −/− mice exhibit higher ethanol and acetaldehyde levels and lower acetate levels in the blood, brain, and liver than Aldh2 +/+ mice [57, 58]. Further, they are more susceptible to ethanol-induced body weight loss [59], but show no change in mortality [60]. Aldh2 −/− mice are more sensitive to the toxic effects of inhaled acetaldehyde [61] and exhibit more frequent mutations in the T cell receptor site than their corresponding wild-type [62]. A single oral dose of ethanol in Aldh2 −/− downregulates the alcohol-metabolizing CYP2E1 mRNA [63], which suggests that there is compensation due to an abundance of acetaldehyde. This treatment has also been shown to decrease hepatic malondialdehyde and increase hepatic glutathione, both markers of oxidative stress, in Aldh2 −/− mice [64]. Acetaldehyde adducts are also increased in Aldh2 −/− mice. These mice have been used to determine ethanol- and acetaldehyde-induced cholinergic changes in the hippocampus. The null mice exhibit decreases in choline acetyltransferase mRNA and protein; however, neurotrophins (nerve growth factor or brain-derived neurotrophic factor) remain unaffected [65], indicating that aldehydes have a selective effect in the brain. Aldh2 −/− mice also exhibit alcohol avoidance in a test of preference and difference in liver or brain acetaldehyde levels [55]. ALDH2 also appears to influence bone growth and cardiac function, as demonstrated by reductions in trabecular bone formation and cardiomyocyte function in Aldh2 −/− mice treated with alcohol [66, 67]. Stomach DNA adducts are dramatically increased after chronic ethanol feeding of Aldh2 −/− mice [68, 69] and acute ethanol treatment increases hepatic oxidative DNA adducts in null mice [70, 71]. The Aldh2 −/− strain represents a valuable strain that can be used to identify functions of ALDH2 in ethanol metabolism and toxicity.
ALDH2 conditional knockout: A “knockout-first” conditional allele for Aldh2 has been developed by Skarnes and colleagues [72]. These mice have been crossed with FLP mice to generate Aldh2 floxed conditional knockout (Aldh2 f/f ) mice, which can be further crossed with specific CRE mouse lines to generate cell-specific Aldh2 knockout mice. As expected, Aldh2 f/f mice develop normally and exhibit no observed phenotype (unpublished observation). To date, no ethanol studies have been conducted in these mice. This strain can be used to study tissue-specific contributions of ALDH2 in ethanol metabolism and toxicity.
ALDH1B1 global knockout: The Aldh1b1 −/− strain was recently generated by Vasiliou and coworkers (Singh S, Vasiliou V et al., manuscript in preparation) (Table 22.1). The Aldh1b1 −/− mice develop normally and show no overt phenotype. In agreement with the catalytic properties of ALDH1B1 (i.e., the second lowest Km for acetaldehyde oxidation [73]), these mice exhibit higher blood concentrations of acetaldehyde following acute ethanol administration [manuscript in preparation]. The Aldh1b1 −/− strain represents the first animal model for the study of ALDH1B1 enzyme in ethanol-induced tissue injury.
ALDH1A1 global knockout: The Aldh1a1 −/− mouse line has been generated by Fan et al. [74] (Table 22.1). These mice are fertile and exhibit no overt phenotype, except that aged Aldh1a1 −/− mice display ~2.4-fold higher cataract incidence than wild-type mice [75]. While ALDH1A1 primarily metabolizes retinaldehyde, it also plays a role in acetaldehyde metabolism. Genetic variants of ALDH1A1 (that result in low enzyme activity) have been associated with increased alcohol sensitivity in Caucasians [29]. Therefore, the Aldh1a1 −/− mouse line represents a useful animal model for investigation of the ALDH1A1 enzyme in ethanol toxicities.
22.7 Mouse Models with GSH Deficiency
GCLC conditional (Gclc f/f ) knockout: The global gene knockout of Gclc results in embryonic lethality, indicating an essential role of GSH in mouse development [76]. The Gclc f/f strain was developed and originally characterized by Chen and colleagues [77]. The in vivo role of hepatic GSH has been investigated using the hepatocyte-specific Gclc knockout (Gclc h/h ) mice created by intercrossing Gclc f/f and Alb-Cre mice [77]. Gclc h/h mice experience almost complete loss of hepatic GSH (~5 % of normal) and die from acute liver failure when mitochondrial failure occurs [77]. Chronic administration of N-acetylcysteine, a treatment that promotes only a mild increase in liver GSH levels (to 8 % of normal) but partially preserves mitochondrial function, allows Gclc h/h mice to survive to adulthood, albeit with the serious liver pathologies fibrosis and cirrhosis [78]. These studies demonstrate an essential role of GSH in normal functioning of the liver. The Gclc f/f mice represent a unique model that can be used to elucidate cell-specific functions of GSH in ethanol metabolism and toxicity.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree