21 The Use of Molecular Profiles in the Management of Breast Cancer
Introduction
Biomarkers
Biomarkers are verified when their presence has been confirmed by more than one methodology. Validation of an assay, however, requires more stringency. A test is said to be validated when it is reproducible in the initial laboratory developing the assay, when it has been demonstrated to correlate with outcomes in an independently derived dataset with no overall relation to the original discovery set, and when it has been tested for its correlation to outcomes (either prognostic or predictive) in the population of patients, tumors, and treatments for the question being addressed. For example, the OncotypeDx assay has been extensively validated for a relatively narrow and homogeneous subset of breast cancers (ER-positive, nodenegative, and largely HER2-negative), providing an assessment of residual risk of distant relapse after assuming the patient had taken 5 years of tamoxifen. This test can therefore be used to guide treatment choices in this subset of patients, but not in those with a different subtype of breast cancer.
Tumor tissue is most often queried with immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). Traditional markers by IHC include ER, PR, HER2, Ki67 or MIB-1 (markers of proliferation), and p63 (to determine invasiveness if uncertain) (Fig. 21-1). HER2 is the only marker currently ascertained routinely by FISH, although topoisomerase II amplification in HER2-positive breast cancer may become useful if validated (Fig. 21-2). IHC and FISH have the advantages of being cheap, reproducible in multiple laboratories, and able to demonstrate tumor heterogeneity. They also have the most long-term clinical usage and are well validated. IHC typically measures proteins (presumably the business end of gene expression), and FISH measures gene copy number (a more remote endpoint relative to protein function). Disadvantages of these techniques are that quality assurance among laboratories is poor or fair, they can interrogate only a few molecular pathways, and the intensity of IHC at least is linear only over a small range. Newer techniques use RT-PCR (reverse transcriptase polymerase chain reaction) or cDNA arrays to measure mRNA levels of various genes.
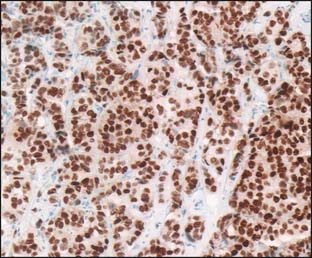
Figure 21-1 Immunohistochemistry for estrogen receptor.
(From www.nibib.nih.gov/nibib/Image/Eadvances/scannerERbiopsy2.jpg.)
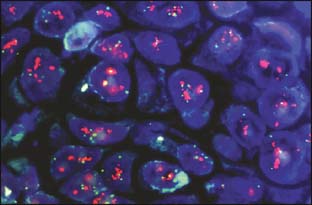
Figure 21-2 Fluorescence in situ hybridization for HER2/neu overamplification.
(From Digital Atlas of Breast Pathology, Meenakshi Singh, MD, www.hsc.stonybrook.edu/gyn-atlas/breast-atlas/atlasrcp.htm)
Prognostic Profiles
The Intrinsic Subtype Assay
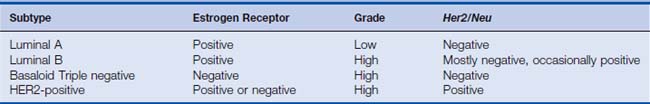
As discussed in Chapter 19, Perou and colleagues1–3 first described molecular profiles of subtypes of breast cancer, generally concordant with clinical thinking. The intrinsic subtype assay requires fresh frozen tissue, although newer assays depending on RT-PCR or antibody panels in fixed tissues are being developed (Table 21-1). Luminal A assays are ER-positive, generally low grade with low proliferative thrust, and HER2-negative. Luminal B assays are ERpositive, high grade with high proliferative thrust, and occasionally HER2-positive. Frequently, they may be PR-low. HER2-positive breast cancers are driven by the characteristic HER2 gene amplification and are biologically aggressive with high grade and proliferation indices. These may be ER-negative or -positive. Basaloid or triple-negative breast cancers are characterized by the lack of HER2, ER, and PR, but are heterogeneous in other respects. They have high grade and proliferation. Sub-subtypes can be identified, although to date, none of these patterns is validated nor carries clear therapeutic implications. This represents a fertile area for target and drug discovery. It is possible that a gene signature associated with BRCA1 dysfunction may become an important subtype given the provocative data generated in the PARP inhibitor trials.
The OncotypeDx Recurrence Score
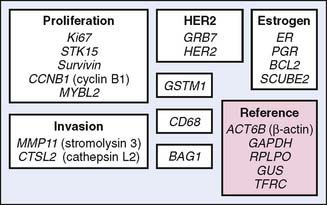
The OncotypeDx assay (Genomic Health, Inc, Redwood City, California) is a multiplex RT-PCR measurement of mRNA levels for 21 genes: 16 breast cancer–related and five housekeeping genes to normalize the data (Fig. 21-3). Fixed tissue is used. The expression levels of these 16 cancer genes were placed into an algorithm correlated with distant disease-free survival at 10 years in 447 patients. This assay was subsequently validated in ER-positive node-negative breast cancer tissues obtained from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14 and B-20 trials.4,5 The recurrence score (RS) as a continuous variable correlates with the distant disease-free survival (DDFS) at 10 years, assuming that the patient has taken tamoxifen for 5 years (Fig. 21-4). More recent work suggests that the RS also will correlate with outcome in node-positive breast cancers.6,7 As highlighted in Chapter 19, this assay is being prospectively validated in the TAILORx trial (Trial Assigning IndividuaLized Options for Treatment) and is used extensively in clinical decision making.8
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree