Tomohiro Nakamura, Louis R. Lapierre, Malene Hansen, Stuart A. Lipton
The Neurobiology of Aging: Free Radical Stress and Metabolic Pathways
Environmental stressors and several genetic pathways play complex and crucial roles in the neurobiology and control of aging. This chapter will summarize current knowledge on these two specific research areas divided into two sections, one on free radical stressors and the other on genetic control of metabolic pathways.
Nitrosative and Oxidative Stress in the Neurobiology of Aging
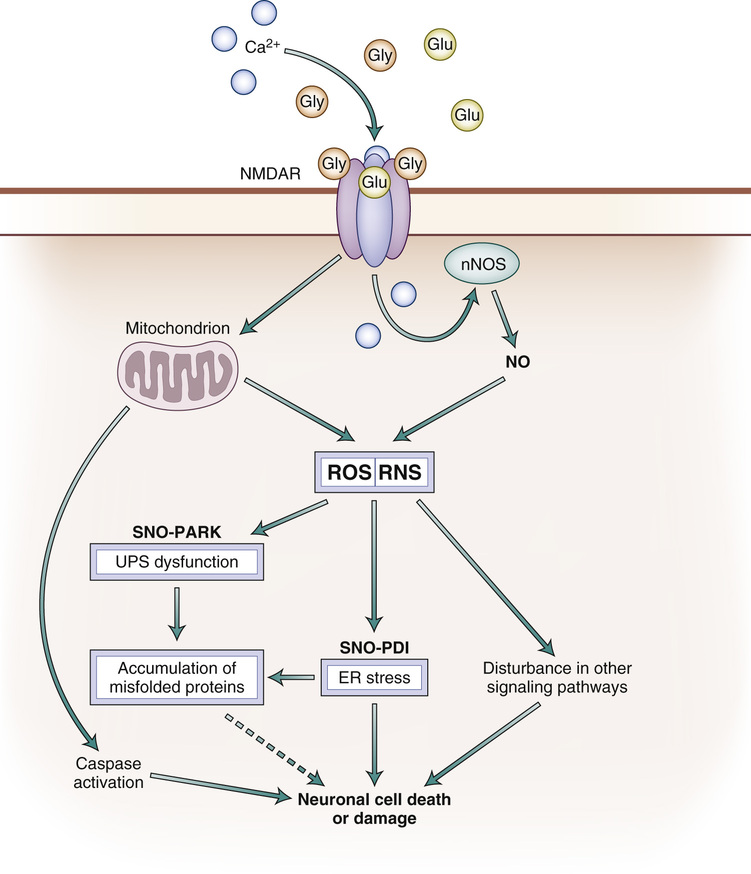
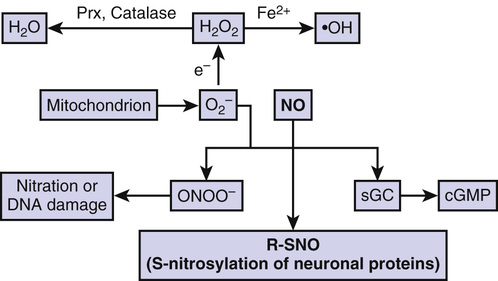
Aging represents a major risk factor for neurodegenerative diseases, such as Parkinson disease (PD), Alzheimer disease (AD), amyotrophic lateral sclerosis (ALS), polyglutamine (polyQ) diseases such as Huntington disease (HD), glaucoma, human immunodeficiency virus (HIV)–associated neurocognitive disorder (HAND), multiple sclerosis, and ischemic brain injury, to name but a few.1–5 Although many intra- and extracellular molecules may participate in neuronal injury and loss, the accumulation of nitrosative and oxidative stress, due to excessive generation of reactive nitrogen species (RNS) such as nitric oxide (NO) and of reactive oxygen species (ROS), appears to be a potential factor contributing to neuronal cell damage and death.6,7 A well-established model for NO production entails a central role of the N-methyl-D-aspartate (NMDA)–type glutamate receptors in the nervous system. Excessive activation of NMDA receptors drives Ca2+ influx, which in turn activates neuronal NO synthase (nNOS) as well as the generation of ROS (Fig. 11-1).8,9 Accumulating evidence suggests that NO can mediate protective and neurotoxic effects by reacting with cysteine residues of target proteins to form S-nitrosothiols (SNOs), a process termed S-nitrosylation because of its effects on the chemical biology of protein function. Importantly, normal mitochondrial respiration also generates free radicals, principally ROS, and one such molecule, superoxide anion (O2−), reacts rapidly with free radical NO under nitrosative stress conditions to form the very toxic product peroxynitrite (ONOO−) (Fig. 11-2).10,11
An additional feature of most neurodegenerative diseases is accumulation of misfolded and/or aggregated proteins.12–15 These protein aggregates can be cytosolic, nuclear, or extracellular. Importantly, protein aggregation can result from a mutation in the disease-related gene encoding the protein, or posttranslational changes to the protein engendered by nitrosative and oxidative stress.16 A key theme of this chapter, therefore, is the hypothesis that age-related nitrosative or oxidative stress contributes to protein misfolding in the brains of neurodegenerative patients. In the first section of this chapter, we discuss specific examples showing that S-nitrosylation of ubiquitin E3 ligases such as parkin or endoplasmic reticulum (ER) chaperones such as protein disulfide isomerase (PDI) are critical factors for the accumulation of misfolded proteins in neurodegenerative diseases such as PD and other conditions.17–20
Protein Misfolding in Neurodegenerative Diseases
A shared histologic feature of many neurodegenerative diseases is the accumulation of misfolded proteins that adversely affect neuronal connectivity and plasticity and trigger cell death signaling pathways.12,15 For example, degenerating brains contain aberrant accumulations of misfolded aggregated proteins, such as α-synuclein and synphilin-1 in PD, and amyloid-β (Aβ) and tau in AD. The inclusions observed in PD are called Lewy bodies (LBs) and are mostly found in the cytoplasm. AD brains show intracellular neurofibrillary tangles, which contain tau, and extracellular plaques, which contain Aβ. Other disorders manifesting protein aggregation include HD (polyQ), ALS, and prion disease.14 These aggregates may consist of oligomeric complexes of non-native secondary structures and demonstrate poor solubility, even in the presence of detergents.
In general, protein aggregates do not accumulate in unstressed healthy neurons due in part to the existence of cellular quality control mechanisms. For example, molecular chaperones are believed to provide a defense mechanism against the toxicity of misfolded proteins because chaperones can prevent inappropriate interactions within and between polypeptides and can promote refolding of proteins that have been misfolded because of cell stress. In addition to the quality control of proteins provided by molecular chaperones, the ubiquitin-proteasome system (UPS) and autophagy-lysosomal degradation are involved in the clearance of abnormal or aberrant proteins.21 When chaperones cannot repair misfolded proteins, they may be tagged via the addition of polyubiquitin chains for degradation by the proteasome. In neurodegenerative conditions, intra- or extracellular protein aggregates are thought to accumulate in the brain as a result of a decrease in molecular chaperone or proteasome activities. In fact, several mutations that disturb the activity of molecular chaperones or UPS-associated enzymes can cause neurodegeneration.15,22,23 Along these lines, postmortem samples from the substantia nigra of PD patients manifest a significant reduction in proteasome activity compared to non-PD controls.24
Historically, lesions that contain aggregated proteins were considered to be pathogenic. Several lines of evidence have suggested that aggregates are formed through a complex multistep process whereby misfolded proteins assemble into inclusion bodies; soluble oligomers of these aberrant proteins are thought to be the most toxic forms via interference with normal cell activities, whereas large insoluble aggregates may be an attempt by the cell to wall off potentially toxic material.25,26
Generation of Reactive Oxygen and Reactive Nitrogen Species
Induction of Ca2+ Influx by NMDA Receptor–Mediated Glutamatergic Signaling Pathways
It is well known that the amino acid glutamate is the major excitatory neurotransmitter in the brain. Glutamate is present in high concentrations in the adult central nervous system and is released for milliseconds from nerve terminals in a Ca2+-dependent manner. After glutamate enters the synaptic cleft, it diffuses across the cleft to interact with its corresponding receptors on the postsynaptic face of an adjacent neuron. Excitatory neurotransmission is necessary for the normal development and plasticity of synapses and for some forms of learning and memory; however, excessive activation of glutamate receptors is implicated in neuronal damage in many neurologic disorders, ranging from acute hypoxic-ischemic brain injury to chronic neurodegenerative diseases. It is currently thought that overstimulation of extrasynaptic NMDA receptors mediates this neuronal damage, whereas, in contrast, synaptic activity may activate survival pathways.27–29 Intense hyperstimulation of excitatory receptors leads to necrotic cell death, but milder or chronic overstimulation can result in apoptotic cell death.30–32
NMDA receptor–coupled channels are highly permeable to Ca2+, thus permitting Ca2+ entry after ligand binding if the cell is depolarized to relieve block of the receptor-associated ion channel by Mg2+.33,34 Subsequent binding of Ca2+ to various intracellular molecules can lead to many significant consequences. In particular, excessive activation of NMDA receptors leads to the production of damaging free radicals (e.g., NO and ROS) and other enzymatic processes, contributing to cell death.*
Ca2+ Influx and Generation of Reactive Oxygen and Reactive Nitrogen Species
Excessive activation of glutamate receptors is implicated in neuronal damage in many neurologic disorders. Olney coined the term excitotoxicity to describe this phenomenon.37,38 This form of toxicity is mediated at least in part by excessive activation of NMDA-type receptors,6,7,39 resulting in excessive Ca2+ influx through a receptor’s associated ion channel.
Increased levels of neuronal Ca2+, in conjunction with the Ca2+-binding protein Ca2+ calmodulin (CaM), trigger the activation of nNOS and subsequent generation of NO from the amino acid L-arginine.8,40 NO is a gaseous free radical (thus highly diffusible) and a key molecule that plays a vital role in normal signal transduction but, in excess, can lead to neuronal cell damage and death. This discrepancy in NO effects on neuronal survival can also be caused by the formation of different NO species or intermediates—NO radical (NO⋅), nitrosonium cation (NO+), nitroxyl anion (NO−, with a high-energy singlet and lower energy triplet forms).11
Studies have further pointed out the potential connection between ROS-RNS and mitochondrial dysfunction in neurodegenerative diseases, especially in PD.5,41 Pesticide and other environmental toxins that inhibit mitochondrial complex I result in oxidative and nitrosative stress, with consequent aberrant protein accumulation.17,18,20,42,43 Administration to animals of complex I inhibitors, such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), 6-hydroxydopamine, rotenone, and paraquat, which result in the overproduction of ROS-RNS, reproduces many of the features of sporadic PD, such as dopaminergic neuron degeneration, upregulation and aggregation of α-synuclein, LB-like intraneuronal inclusions, and behavioral impairment.5,41
Increased nitrosative and oxidative stress are associated with chaperone and proteasomal dysfunction, resulting in the accumulation of misfolded aggregates.16,44 However, until recently, little was known regarding the molecular and pathogenic mechanisms underlying the contribution of NO to the formation of inclusion bodies, such as amyloid plaques in AD or LBs in PD.
Protein S-Nitrosylation and Neuronal Cell Death
Chemical Biology of S-Nitrosylation
Early investigations indicated that NO mediates cellular signaling pathways, which regulate broad aspects of brain function, including synaptic plasticity, normal development, and neuronal cell death.35,45–47 In general, NO exerts physiologic and some pathophysiologic effects via stimulation of guanylate cyclase to form cyclic guanosine-3′,5′-monophosphate (cGMP) or through S-nitros(yl)ation of regulatory protein thiol groups (Fig. 11-2).9,11,44,48–50 S-Nitrosylation is the covalent addition of an NO group to a critical cysteine thiol sulfhydryl (RSH or, more properly, thiolate anion, RS−) to form an S-nitrosothiol derivative (R-SNO). Such modification modulates the function of a broad spectrum of mammalian, plant, and microbial proteins. In general, a consensus motif of amino acids comprised of nucleophilic residues (generally an acid and base) surrounds a critical cysteine, which increases the cysteine sulfhydryl’s susceptibility to S-nitrosylation.51,52 Our group first identified the physiologic relevance of S-nitrosylation by showing that NO and related RNS exert paradoxic effects via redox-based mechanisms. NO is neuroprotective via S-nitrosylation of NMDA receptors (as well as other subsequently discovered targets, including caspases) and yet can also be neurodestructive by the formation of peroxynitrite (or, as later discovered, a reaction with additional molecules such as matrix metalloproteinase-9 [MMP-9] and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]).11,53–60 Examples of SNO proteins known to be present in neurons or brains are listed in Table 11-1. Over the past decade, accumulating evidence has suggested that S-nitrosylation can regulate the biologic activity of a great variety of proteins, in some ways akin to phosphorylation.*
TABLE 11-1
Examples of S-Nitrosylated Proteins Confirmed in Neurons or Brains
| S-Nitrosothiol Targets | Effects of S-Nitrosothiol | Reference |
| Akt | Decreased kinase activity Increased cell death | 210, 211 |
| Caspases | Decreased activity Suppression of cell death | 56, 54, 212, 55 |
| Cdk5 | Activation of kinase activity Augmentation of cell death | 213–215 |
| Dexras1 | Activation of GTPase Regulation of iron homeostasis | 216, 217 |
| Drp1 | Excessive mitochondrial fission Synaptic damage | 218–220 |
| NSF | Enhanced interaction with GluR2 Regulation of exocytosis | 221, 222 |
| GAPDH | Enhanced interaction with Siah1 Activation of p300 and CBP Augmentation of cell death | 60, 223, 224 |
| MAP1B | Enhanced interaction with microtubules Axon retraction | 225 |
| MMP-9 | Activation Augmentation of cell death | 59 |
| NMDAR (NR1 and NR2A) | Inhibition Suppression of cell death | 11, 58 |
| Parkin | Decrease in E3 ligase activity Augmentation of cell death | 17, 18 |
| PDI | Decreased activity Accumulation of misfolded proteins Augmentation of cell death | 20, 121–123 |
| Prx2 | Decreased peroxidase activity Augmentation of cell death | 226 |
| PTEN | Decrease in phosphatase activity Augmentation of cell survival | 227, 228 |
| XIAP | Decrease in E3 ligase activity Augmentation of cell death | 229, 230 |
Chemically, NO is often a good “leaving group,” facilitating further oxidation of a critical thiol to a disulfide bond between neighboring (vicinal) cysteine residue or, via reaction with ROS, to sulfenic (SOH), sulfinic (SO2H), or sulfonic (SO3H) acid derivatization of the protein, as occurs, for example, on the enzyme MMP-9.18,20,59,68 Alternatively, S-nitrosylation may possibly produce a nitroxyl disulfide, in which the NO group is shared by vicinal cysteine thiols.69
In addition, S-nitrosylation has been reported to influence other oxidative posttranslational modifications of cysteine residues. For example, S-nitrosylated as well as sulfenated cysteine residues may react with glutathione, producing S-glutathionylated proteins.70 Moreover, S-sulfhydration (modification of a cysteine residue by hydrogen sulfide [H2S]) generally occurs on the same cysteine residue(s) that can also undergo S-nitrosylation,71,72 suggesting that S-nitrosylation may potentially promote the formation of S-sulfhydration.73 Further studies are needed to reveal the chemical relationship between S-nitrosylation and other types of posttranslational modifications of thiol. Protein S-nitrosylation and other reactions with cysteine sulfhydryls should not be confused with protein nitration, in which NO is also implicated often via peroxynitrite (ONOO−) reacting with a tyrosine residue to generate a nitro-tyrosine adduct.
Analyses of mice deficient in nNOS or iNOS (inducible NOS) have confirmed that NO is an important mediator of cell injury and death after excitotoxic stimulation; NO generated from nNOS or iNOS can be detrimental to neuronal survival.74,75 In addition, inhibition of NOS activity ameliorates the progression of disease pathology in animal models of PD, AD, and ALS, suggesting that excess generation of NO plays a pivotal role in the pathogenesis of several neurodegenerative diseases.76–79 Although the involvement of NO in neurodegeneration has been widely accepted, the chemical relationship between nitrosative stress and accumulation of misfolded proteins has remained obscure. Some studies, however, have shed light on molecular events underlying this relationship. Specifically, we recently presented physiologic and chemical evidence that S-nitrosylation modulates the ubiquitin E3 ligase activity of parkin,17–19 and chaperone and isomerase activities of PDI,20 contributing to protein misfolding and neurotoxicity in models of neurodegenerative disorders.
S-Nitrosylation and Parkin
Identification of errors in the genes encoding parkin (an ubiquitin E3 ligase) and ubiquitin carboxyl-terminal hydrolase L1 (UCH-L1) in rare familial forms of PD has implicated possible dysfunction of the UPS in the pathogenesis of sporadic PD as well. The UPS represents an important mechanism for proteolysis in mammalian cells. Formation of polyubiquitin chains constitutes the signal for proteasomal attack and degradation. An isopeptide bond covalently attaches the C-terminus of the first ubiquitin in a polyubiqutin chain to a lysine residue in the target protein. The cascade of activating (E1)-, conjugating (E2)-, and ubiquitin-ligating (E3)–type enzymes catalyzes the conjugation of the ubiquitin chain to proteins. In addition, individual E3 ubiquitin ligases play a key role in the recognition of specific substrates.80
PD is the second most prevalent neurodegenerative disease and is characterized by the progressive loss of dopamine neurons in the substantia nigra pars compacta. The appearance of LBs that contain misfolded and ubiquitinated proteins generally accompanies the loss of dopaminergic neurons in the PD brain. Such ubiquitinated inclusion bodies are the hallmark of many neurodegenerative disorders. Age-associated defects in intracellular proteolysis of misfolded or aberrant proteins might lead to the accumulation and ultimately deposition of aggregates within neurons or glial cells. Although such aberrant protein accumulation had been observed in patients with genetically encoded mutant proteins, evidence from our laboratory suggests that nitrosative and oxidative stress are potential causal factors for protein accumulation in the much more common sporadic form of PD. As illustrated later, nitrosative and oxidative stress, commonly found during normal aging, can mimic rare genetic causes of disorders, such as PD, by promoting protein misfolding in the absence of a genetic mutation.17–19 For example, S-nitrosylation and further oxidation (e.g., sulfonation) of parkin result in dysfunction of this enzyme and thus of the UPS.17,18,81–85 We and others have found that nitrosative stress triggers S-nitrosylation of parkin (forming SNO-parkin), not only in rodent models of PD but also in the brains of human patients with PD and the related α-synucleinopathy DLBD (diffuse LB disease). Parkin has multiple cysteine residues that can react with NO to form SNO-parkin. S-Nitrosylation of parkin initially stimulates ubiquitin E3 ligase enzymatic activity, followed by a decrease in enzyme activity, producing a futile cycle of dysfunctional E3 ligase–UPS stimulation.18,19,86 We also found that the pesticide rotenone led to the generation of SNO-parkin and thus consequent inhibition of ubiquitin E3 ligase activity. Moreover, S-nitrosylation–induced inactivation of E3 ligase activity is also associated with a decreased neuroprotective effect of parkin.17 Along these lines, SNO-parkin is increased in PD patients’ brains, suggesting that long-term S-nitrosylation of parkin may be pathogenic.17–19 Nitrosative and oxidative stress can also alter the solubility of parkin via posttranslational modification of cysteine residues, which may concomitantly compromise its protective function.85,87–89
In contrast, S-sulfhydration of parkin activates its enzymatic activity and its neuroprotective effect.72 Mass spectrometry analysis was used to identify five cysteine residues (Cys59, -95, -182, -212, and -377) as S-sulfhydration sites, among which Cys95 appeared to be the principal site of S-sulfhydration. Interestingly, Cys95 also serves as a target for S-nitrosylation.72 Moreover, S-sulfhydration of parkin is markedly decreased in PD brains, consistent with the notion that parkin sulfhydration has beneficial effects. Thus, although further studies will be needed to reveal the relationship of S-nitrosylation and S-sulfhydration in individual proteins, at least in the case of parkin, it appears that the two gasotransmitters (i.e., NO and H2S) may influence PD pathogenesis via cysteine modifications.
In addition to rare mutations in parkin causing familial PD, mutations in the gene PINK1 are also associated with hereditary PD cases. Emerging evidence points to the possibility that parkin, together with PINK1, participates in mitophagy, whereby damaged mitochondria are removed by the cellular recycling process of autophagy (see next section).90 In the proposed model, PINK1 is initially translocated to impaired mitochondria, which in turn recruit parkin from the cytosol to the damaged mitochondrial membrane. Recent evidence suggests that PINK1-phosphorylated mitofusin 2 may be a receptor for parkin on the mitochondrial membrane.91 Parkin then ubiquitinates mitochondrial outer membrane proteins to enhance the autophagic removal of the unhealthy mitochondria. New evidence suggests that SNO-parkin can promote the removal of damaged mitochondria.92 That report showed that S-nitrosylation of Cys323 transiently activates its E3 ligase activity, facilitating the mitochondrial degradation process. This finding raises the interesting hypothesis that nitrosative stress initially induces S-nitrosylation of parkin at Cys323, resulting in a short-lived increase in its E3 ligase activity. In turn, this activation of parkin mediates a neuroprotective effect of NO via promotion of mitophagy and hence disposal of unhealthy mitochondria. In contrast, pathologically prolonged generation of NO would S-nitrosylate additional cysteine residues in parkin, inhibiting its E3 ligase and neuroprotective activities. Further work will be needed, however, to elucidate more clearly these opposing effects of SNO-parkin on neuronal survival.
In addition to its E3 ligase activity, parkin also suppresses the transcription of the oncogene p53, contributing to its neuroprotective action against PD-associated apoptosis of dopaminergic neurons.93 We recently reported that S-nitrosylation of parkin decreases its activity as a transcriptional repressor of p53, leading to the upregulation of p53 expression and thus neuronal cell death.94 Consistent with this notion, in postmortem human PD brains, the levels of SNO-parkin and p53 are increased in a correlative manner. Hence, S-nitrosylation appears to affect both the ubiquitin E3 ligase and transcriptional repressor activities of parkin, coordinately contributing to the pathogenesis of sporadic PD.
S-Nitrosylation of Protein Disulfide Isomerase
This mediates protein misfolding and neurotoxicity in cell models of PD and AD. The ER normally participates in protein processing and folding but undergoes a stress response when immature or misfolded proteins accumulate.95–98 ER stress stimulates two critical intracellular responses. The first represents expression of chaperones that prevent protein aggregation via the unfolded protein response (UPR), and is implicated in protein refolding, posttranslational assembly of protein complexes, and protein degradation. This response is believed to contribute to adaptation during altered environmental conditions, promoting maintenance of cellular homeostasis. The second ER stress response, termed ER-associated degradation (ERAD), specifically recognizes terminally misfolded proteins for retrotranslocation across the ER membrane to the cytosol, where they can be degraded by the UPS. Additionally, although severe ER stress can induce apoptosis, the ER withstands relatively mild insults via expression of stress proteins such as glucose-regulated protein (GRP) and PDI, which ameliorate the accumulation of misfolded proteins. These proteins behave as molecular chaperones that assist in the maturation, transport, and folding of secretory proteins.
During protein folding in the ER, PDI (designated PDIA1) can introduce disulfide bonds into proteins (oxidation), break disulfide bonds (reduction), and catalyze thiol-disulfide exchange (isomerization), thus facilitating disulfide bond formation, rearrangement reactions, and structural stability.99 PDI has four domains that are homologous to thioredoxin (TRX; a, b, b′, and a′). Only two of the four TRX-like domains (a and a′) contain a characteristic redox-active CXXC motif, and these two thiol-disulfide centers function as independent active sites.100–103 Several mammalian PDI homologues, such as ERp57 (PDIA3) and PDIp (PDIA2), also localize to the ER and may manifest similar functions.104,105 Increased expression of PDIp in neuronal cells under conditions mimicking PD suggest the possible contribution of PDIp to neuronal survival.104
In many neurodegenerative disorders and cerebral ischemia, the accumulation of immature and denatured proteins results in ER dysfunction,104,106–108 but upregulation of PDI represents an adaptive response promoting protein refolding and may offer neuronal cell protection.104,105,109,110 In addition, it is generally accepted that excessive generation of NO can contribute to activation of the ER stress pathway, at least in some cell types111,112 Molecular mechanisms whereby NO induces protein misfolding and ER stress, however, have remained enigmatic until recently. The ER normally manifests a relatively positive redox potential in contrast to the highly reducing environment of the cytosol and mitochondria. This redox environment can influence the stability of protein S-nitrosylation and oxidation reactions.113 Interestingly, we previously reported that excessive NO can lead to S-nitrosylation of the active site thiol groups of PDI, and this reaction inhibits its isomerase and chaperone activities.20 Mitochondrial complex I insult by rotenone can also result in S-nitrosylation of PDI in cell culture models. Moreover, we found that PDI is S-nitrosylated in the brains of virtually all cases examined of sporadic AD and PD. Under pathologic conditions, it is possible that both cysteine sulfhydryl groups in the TRX-like domains of PDI form S-nitrosothiols. Unlike formation of a single S-nitrosothiol, which is commonly seen after denitrosylation reactions catalyzed by PDI,63 dual nitrosylation may be relatively more stable and prevent subsequent disulfide formation on PDI. Therefore, we speculate that these pathologic S-nitrosylation reactions on PDI are more easily detected during neurodegenerative conditions.
Additionally, it is possible that vicinal (nearby) cysteine thiols reacting with NO can form nitroxyl disulfide,69 and such a reaction may potentially occur in the catalytic site of PDI to inhibit enzymatic activity. To determine the consequences of S-nitrosylated PDI (SNO-PDI) formation in neurons, we exposed cultured cerebrocortical neurons to neurotoxic concentrations of NMDA, thus inducing excessive Ca2+ influx and consequent NO production from nNOS. Under these conditions, we found that PDI was S-nitrosylated in an NOS-dependent manner. SNO-PDI formation led to the accumulation of polyubiquitinated misfolded proteins (e.g., synphilin-1 and α-synuclein) and activation of the UPR.20,114–116 Moreover, S-nitrosylation abrogated the inhibitory effect of PDI on the aggregation of proteins observed in LB inclusions.20,117 Other studies have also demonstrated that in AD, PDI accumulates in neurofibrillary tangles, which contain hyperphosphorylated aggregated tau protein, suggesting that SNO-PDI may also contribute to the accumulation of aggregated tau.118,119 S-Nitrosylation of PDI also prevented its attenuation of neuronal cell death triggered by ER stress, misfolded proteins, or proteasome inhibition. Further evidence has suggested that SNO-PDI may in effect transport NO to the extracellular space, where it could conceivably exert additional adverse effects.63 In addition, S-nitrosylation of PDI at its active cysteine sites may facilitate S-glutathionylation of this protein.120 Thus, the potential role of S-glutathionylated PDI in neurodegeneration needs to be elucidated in future studies.
Similar to the finding that SNO-PDI contributes to the pathogenesis of AD and PD, SNO-PDI may also exacerbate pathologic conditions such as ALS, stroke, prion disease, and sleep disorders. For example, in cellular models of ALS, S-nitrosylation of PDI increases aggregation of the G93A mutant of superoxide dismutase 1 (mutSOD1), which is linked to a familial form of ALS, and increases neuronal cell death.121–123 Also, S-nitrosylation of PDI in a stroke model contributes to the accumulation of ubiquitinated protein aggregates.124 Moreover, S-nitrosylation of PDI mediates selective degeneration of hypothalamic orexin-containing neurons in a model of sleep disorders.125 Finally, consistent with its pathologic role, substantial levels of SNO-PDI are present in the spinal cords of mutSOD1 mice and human patients with sporadic ALS, in brains of an animal model of prion disease, and in the hypothalamus of sleep-deprived mice.121–123,125,126
Next, rather than neurodegenerative disorders, we consider the normal aging brain. The quality control machinery for protein expression, composed of molecular chaperones, the UPS, and the autophagy-lysosome system, have been reported to be impaired in the aging brain.21,127 Additionally, inclusion bodies similar to those found in neurodegenerative disorders can appear in brains of normal aged individuals as well as in those with subclinical manifestations of disease.128 However, we have not found detectable quantities of SNO-parkin or SNO-PDI in the normal aged brain.17,18,20 Hence, we speculate that S-nitrosylation of these proteins and others may further disrupt the quality control machinery, thus contributing to the susceptibility of the aging brain to neurodegenerative conditions.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree