Fig. 18.1
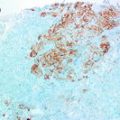
This four-panel set of photomicrograph shows the histology displayed in sections of 4 distinct male breast cancers and demonstrates morphological heterogeneity in these tumours. All ×400
Gene expression studies have confirmed that there is a great deal of heterogeneity amongst FBC, and there is evidence that immunohistochemistry surrogates can be used to classify FBCs into clinically-relevant different molecular subgroups [7]. Some studies have applied these immunohistochemistry surrogates directly to cohorts of MBC the findings of which are displayed in Table 18.1 [5, 6, 8, 9]. As is seen when looking at similar studies in FBC, comparison across the different studies is difficult as classifications vary for each study. In general though, a luminal A-like subtype (estrogen receptor (ER) and/or progesterone receptor (PR) positive, human epidermal growth factor receptor 2 (HER-2) negative) is the most common classification seen in MBC. With regards to immunohistochemical markers and survival, PR negativity and p53 accumulation have been reported to be associated with decreased survival in MBC (3), as has overexpression of the proliferation markers cyclin A and cyclin B [7]. Androgen receptor (AR) expression in luminal A MBC has also been shown to be associated with improved overall survival compared to matched FBC [6]. Using hierarchical clustering, it was also shown that AR clustered with estrogen receptor beta (ERβ) in MBC while in FBC, estrogen receptor alpha (ERα) and PR clustered, which could indicate a difference in the biology of breast cancer between the genders (Fig. 18.2).
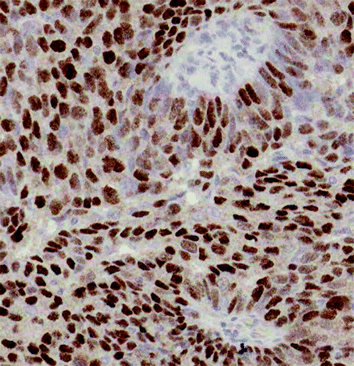
Table 18.1
Studies in which male breast cancers have been categorised into the molecular subgroups defined in FBC
Reference | Molecular subtype | IHC classification used | % |
|---|---|---|---|
[8] n = 130 | Luminal A | ER and/or PR positive; Her2 negative; Ki67 low | 75 |
Luminal B | ER and/or PR positive; Her2 positive and/or Ki67 high | 21 | |
Her2-driven | ER and PR negative; Her2 positive | 0 | |
Basal-like | ER, PR and Her2 negative; CK5/6, CK14 and/or EGFR positive | 3 | |
Unclassifiable | Negative for all 6 markers | 1 | |
[9] n = 183 | Luminal A | ER and/or PR positive; Her2 negative | 81 |
Luminal B | ER and/or PR positive; Her2 positive | 11 | |
Her2-like | ER and PR negative; Her2 positive | 0 | |
Core-basal | ER, PR and Her2 negative; CK5/6 and/or EGFR positive | 1 | |
5-negative profile | ER, PR and Her2 negative; CK5/6 and EGFR negative | 0 | |
[5] n = 189 | Luminal A | ER and/or PR positive; Her2 negative | 68 |
Luminal B | ER and/or PR positive; Her2 positive | 26 | |
Her2-positive | ER and PR negative; Her2 positive | 2 | |
Triple negative | ER and PR negative; Her2 negative | 4 | |
[6] n = 203 | Luminal A | ER and/or PR positive; Her2 negative | 98 |
Luminal B | ER and/or PR positive; Her2 positive | 0 | |
Her2-positive | ER and PR negative; Her2 positive | 2 | |
Basal | ER and PR negative; Her2 negative; CK5/6 positive | 0 |

Fig. 18.2
Part of a male breast cancer showing strong nuclear staining with an antibody to the androgen receptor gene (AR). AR appears to be associated with better survival in MBC. Hierarchical clustering shows that AR clusters with estrogen receptor beta (ERβ) in MBC whereas in FBC, estrogen receptor alpha (ERα) and PR cluster
DNA Studies
Some studies have attempted to determine specific genomic abnormalities present in MBC and relate these to prognosis and/or compare them to FBC. One of the first of these studies used comparative genomic hybridisation (CGH) of 39 MBC and showed gains of 1q, 8q, 16p, 17q, Xq, 20q and Xp and losses of 8p, 16q, 13q, 6q, 11q and 22q [9]. This pattern was common to genomic imbalances observed in FBC. Another study looked at a cohort of 106 MBCs analysed by multiplex ligation-dependent probe amplification analyses to determine relative gene copy numbers for specific breast cancer genes [10]. A distinctive group of cancers with a poor prognosis were identified, characterized by frequent copy number gains in 6 genes. Amplification of one of these genes, CCND1, was also found to be an independent prognostic factor. Interestingly, gains in some of these genes were not seen in FBC cases. Another study of 56 MBC cases found that compared to FBC, MBC was more likely to have genomic gains and less likely to have genomic losses [11]. Hierarchical clustering revealed two distinct genomic subgroups, termed male-simple and male-complex, one of which (male-simple) appeared distinct from the six subgroups identified in the FBC reference cohort. Finally, a recently published genome-wide association study (GWAS) of 823 MBC cases identified a single nucleotide polymorphism (SNP) at 14q24.1 in the RAD51B gene that was significantly associated with MBC risk as was TOX3 (16q12.1) [12].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree