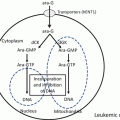
Fig. 12.1
The molecular mechanism of action of ATO in APL. ATO binds to the B2 domain of the PML moiety of the PML-RARα protein, inducing binding of UBC9 followed by multimerization and SUMOylation of PML. SUMOylated PML-RARα fusion protein is brought to the PML moiety and further poly-ubiquitinated, finally leading to degradation by the proteasome. ATO treatment leads to removal of the corepressor complex, in which PML-RARα binds to DNA. The coactivator complex then binds to DNA, and this event leads to granulocyte differentiation and cell death of APL cells
12.5 Resistance to ATO in APL
During the use of ATO as a front therapy for APL, it has demonstrated that some patients develop and acquire ATO resistance. However, the mechanisms underlying this phenomenon remain unknown compared with that of ATRA resistance in APL. As mentioned above, ATO binds directly to the cysteine motif (C212/C213) in the PML-B2 domain of the PML-RARα fusion protein, resulting in the SUMOylation, multimerization, and degradation [40]. This would suggest that the PML-B2 domain is a critical mediator ATO’s chemotherapeutic effect, and mutations affecting this domain may contribute to the development of ATO-resistant APL. Indeed, mutations have been identified in the PML-B2 domain (A216V, L218P, S214L, A216T, S220G) in case of clinically acquired ATO-resistant APL, consistent with the existence of a mutational hotspot (S214-S220) within the PML-RARα fusion protein. Mutations within the putative hotspot resulted in the impairment of direct ATO binding to PML-RARα due to conformational changes in the ATO binding sites [46, 47]. These results suggest that the existence of a mutational hotspot (S214-S220) within the PML-RARα fusion protein in acquired ATO-resistant APL.
More recently, varying responses to ATO linked to different point mutations in the PML moiety of PML-RARα were reported [48]. Among these, the A216, S214L, and A216T mutations attenuated the negative regulation of PML-RARα by ΑΤΟ, resulting in the retention of the fusion protein. In contrast, the L217F and S220G mutations functioned weakly in this context. Interestingly, high-dose ATO, or the combination of ATO and ATRA, can overcome the ATO resistance driven by the acquired point mutations. This may be relevant to the finding that the effects of ATO on APL cells are dose dependent with relatively higher concentrations inducing cell death, while lower concentrations induce partial differentiation of APL cells to mature granulocytes [39]. Ultimately, these data may contribute to improved prognostication and the development of more effective therapeutic strategies for the treatment of APL in the future.
12.6 Conclusion
APL exemplifies the great progress that has been made in the translation of basic research into effective molecularly targeted therapies for hematological malignancies.
Intensive research has addressed many important aspects of APL biology, including transcriptional regulation, nuclear organization, epigenetics, and the role of proteolysis in leukemogenesis, contributing to the development of new therapeutic strategies with significant clinical impact. APL is the first hematological malignancy to be cured using molecular-targeting therapy with ATRA and ATO, and the details of the molecular mechanisms underlying the curative effects of ATO, predominantly mediated by the degradation of the PML-RARα oncoprotein, are gradually being clarified. Future studies will aim to develop sophisticated new therapeutic strategies for the treatment of APL based on a more comprehensive understanding of the molecular mechanisms involved in the targeting and inhibition of PML-RARα by ATO.
References
1.
2.
3.
4.
5.
Coombs CC, Tavakkoli M, Tallman. Acute promyelocytic leukemia: where did we start, where are we now, and the future. Blood Cancer J. 2015;5:e304. doi:10.1038/ bcj.2015.25.CrossRefPubMedPubMedCentral
6.
Zhu J, Chen Z, Lallemand-Breitenbach V, de The H. How acute promyelocytic leukemia revived arsenic. Nat Rev Cancer. 2002;2:1–9.CrossRef
7.
8.
Miller Jr WH, Schipper HM, Lee JS, Singer J, Waxman S. Mechanisms of arsenic trioxide. Cancer Res. 2001;62:3893–903.
9.
Iland HJ, Seymour JF. Role of arsenic trioxide in acute promyelocytic leukemia. Curr Treat Option Oncol. 2013;14:179–84.CrossRef
10.
11.
12.
Zechel C, Shen XQ, Chambon P, Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 1994;13:1414–24.PubMedPubMedCentral
13.
Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, et al. Regulation of retinoid signaling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–31.CrossRefPubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree