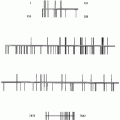
Fig. 1
VU University Medical Centre HPV test algorithm
3.2 The Johns Hopkins Medical Institutions HPV Test Algorithm
This algorithm (Fig. 2) was developed by The Johns Hopkins Medical Institutions, USA (Singhi and Westra 2010; Westra 2014) and was used in the landmark paper by Ang et al. (2010) demonstrating that patients with HPV-related oropharyngeal SCC had better prognosis than patients with HPV-negative disease (82.4 % vs. 57.1 % 3 years survival). The testing strategy incorporates upfront screening with p16 immunohistochemistry (CINtec Histology, Roche mtm laboratories) followed by two tiers of HPV DNA in situ hybridisation. The first tier employs an HPV-16-specific probe, which theoretically would identify the majority of HPV-positive cases (around 95 % of HPV-related oropharyngeal SCCs are HPV-16 positive) and a second tier containing a ‘cocktail’ of HPV probes to detect uncommon oncogenic HPV genotypes (HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -68 DAKO Genpoint). Similar reagents are available from other suppliers: INFORM HPVIII Family 16 probes (Roche Ventana Medical Systems Ltd) are supplied as a cocktail and detect HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -66. Leica Biosystems supply a high-risk HPV probe set (cocktail) that detects HPV-16, -18, -31, -33, -51. Due to licensing restrictions, single genotype-specific probes available in the USA are not for sale in Europe. There is evidence that the testing strategy closely matches the analytical reference test (Schache et al. 2011); furthermore, the clinical importance as a prognostic classifier has been demonstrated in numerous independent cohort studies (Bhatia and Burtness 2015). As a result, the College of American Pathologists and the US National Comprehensive Cancer Network (NCCN) recommend ‘either immunohistochemistry for analysis of p16 expression or HPV in situ hybridisation for detection of HPV DNA in tumour cell nuclei’ for cancer of the oropharynx and the ‘occult primary’ in the head and neck region (College of American Pathologists; National Comprehensive Cancer Network, USA). In the UK the Royal College of Pathologists includes p16 IHC and HPV DNA in situ hybridisation in the datasets for reporting mucosal malignancies of the pharynx (The Royal College of Pathologists, UK). The tests are also being used in clinical trials to identify patients with HPV-positive and HPV-negative SCC (Bhatia and Burtness 2015).
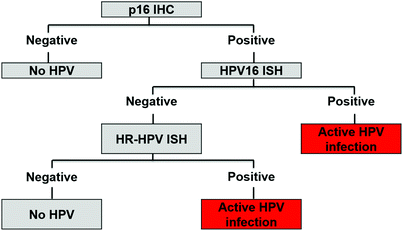

Fig. 2
The Johns Hopkins Medical Institutions HPV test algorithm
One of the inherent problems of using algorithms is that a few cases are inevitably classified in the two indeterminate categories: p16 positive/HPV DNA negative and p16 negative/HPV DNA positive (Table 1). Very few cases are classified in the latter group, which are likely to represent transient HPV infection, without activation of oncogenic effects, or simple interpretative errors. By contrast, the p16-positive/HPV DNA-negative cases represent a dilemma because there is evidence that patients with tumours in this category have similar favourable survival to patients with p16-positive/HPV DNA-positive tumours (Lewis et al. 2010), whereas in other studies this group of patients had unfavourable survival profiles that tracked with the p16-negative/HPV DNA-negative cases (Perrone et al. 2011; Rietbergen et al. 2013b). Interpretation of these data is limited by the small number of patients in the subgroup analysis and further studies are required.
Table 1
Classification of oropharyngeal squamous cell carcinomas using p16 IHC and HPV DNA-specific tests
No. of cases | p16+ HPV DNA+ (%) | p16− HPV DNA− (%) | p16+ HPV DNA− (%) | p16− HPV DNA+ (%) | |
|---|---|---|---|---|---|
Singhi and Westra (2010) | 256 | 71 | 24 | 5 | ND |
Ang et al. (2010) | 315 | 61 | 30 | 7 | 2 |
Lewis et al. (2010) | 239 | 58 | 20 | 20 | 2 |
Thavaraj et al. (2011) | 142 | 53 | 35 | 11 | 1 |
Jordan et al. (2012) | 232 | 60 | 27 | 12 | 2 |
Rietbergen et al. (2013b) | 841 | 19 | 77 | 4 | ND |
4 p16 Immunohistochemistry
CINtec Histology and CINtec Cytology (p16 clone E6H4, Roche mtm laboratories) are the only p16 products that are register as in IVDs, effectively making them the only reagents that can be used for clinical diagnosis. Other antibody clones are available, but are ‘research-use-only’ (RUO) products. CINtec kits are supplied as ‘ready-to-use’ (RTU) products for the Ventana Benchmark autostainer (Roche Ventana Medical Systems Ltd) or as dispensable kits for use on other proprietary automated staining platforms or in manual assays. The assay should be optimised and validated in an appropriately accredited pathology laboratory. Ideally analyte controls should be included on the slides to be tested, and proprietary cell lines controls are available for this purpose (HistoCyte Laboratories Ltd, www.histocyte.com). Alternatively, known positive (e.g. oropharyngeal SCC or cervical intraepithelial neoplasia grade 3) and negative tissue samples can be identified from tissue surplus to diagnostic requirements. Internal positive controls (tissue elements in the sample of interest that show expression of the target molecule) include reticulated tonsil epithelium, which shows patchy moderate staining, and follicular dendritic cells in secondary lymphoid follicles are weakly positive and occasionally fibroblasts showing weak-to-moderate staining. Multinucleated giant cells, if present, are also p16 positive (Schache et al. 2014). Carcinomas that contain oncogenic HPV typically show intense nuclear and cytoplasmic staining in the majority of the malignant cells (Fig. 3a). Westra’s description in the Ang et al. (2010) paper is ‘strong and diffuse nuclear and cytoplasmic staining in 70 % or more of the tumor cells’. Jordan et al. (2012) refined the ‘cut-off’ by comparison with an analytical reference test (RT-PCR for HPV-16, -18, -33 E6/E7 on FFPE tissue) and indicated that the optimum intensity was ≥2 (Scale 0-3), the optimum percentage of tumour cells staining was ≥35 % and the optimum H score (product of intensity and percentage; 0–300) was >60. Accepting a minimum intensity score of 2, at least 30 % of the tumour needed to be positive to show the best correlation with the reference test, and an intensity score of 3 suggests only 20 % of the tumour needs to be p16 positive. In clinical practice, the majority of cases are easily classified in a binary fashion (positive vs. negative), which accounts for the excellent inter-observer agreement for the assay (Thavaraj et al. 2011; Jordan et al. 2012). Clinical trials registering HPV-related oropharyngeal cancers have tended to adopt the >70 % cut-off described above (Bhatia and Burtness 2015). Infrequently, cases with weak staining confined to the cytoplasm of the majority of cells or nuclear staining alone are encountered and are considered to be p16 negative according to the criteria described above. While p16 IHC has features that make interpretation easy, it is recognised to be only an approximation of the HPV status. p16 IHC is considered to be highly sensitive, but lacks specificity, meaning there are occasions when p16 is overexpressed in the absence of HPV infection, using even the most sensitive HPV-specific tests. p16 is an endogenous gene, and increased expression is documented in other tumour types that have no association with HPV infection. With this in mind, there is a case to be made that p16 IHC testing should always be supported by HPV-specific tests (Perrone et al. 2011; Robinson et al. 2012; Rietbergen et al. 2013b), and there is a counter-argument that p16 IHC alone is satisfactory for prognostication and clinical trial recruitment (Ang et al. 2010; Lewis et al. 2010; Bhatia and Burtness 2015).
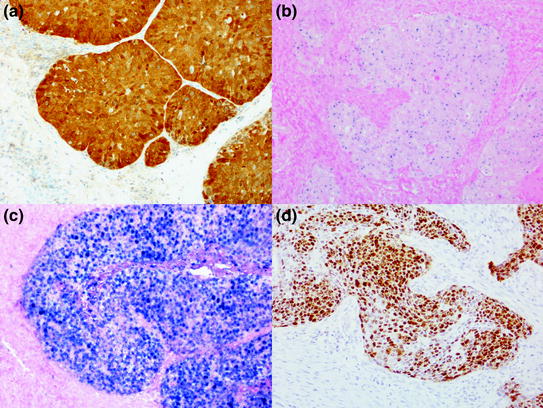

Fig. 3
Photomicrographs showing a typical p16 IHC-positive result (a CINtec histology, Roche mtm laboratories). High-risk HPV DNA ISH (INFORM HPV III Family 16, Roche Ventana Medical Systems Ltd) showing a punctate (b) and a diffuse (c) pattern of staining. High-risk HPV RNA ISH (RNAscope, Advanced Cell Diagnostics) showing brown reaction product in the malignant cells (d)
5 Detection of HPV DNA
Detection of HPV DNA can be achieved by either target amplification (polymerase chain reaction) or signal amplification (in situ hybridisation), but each method has inherent flaws. Non-quantitative PCR techniques tend to be too sensitive (producing false positive results), which can be ameliorated by employing quantitative PCR techniques (Schache et al. 2011). Assessment of PCR products on gels can be subjective, and adjusting detection thresholds in quantitative PCR determines the sensitivity and specificity of the assay. DNA in situ hybridisation is characterised by high specificity, but limited sensitivity, the limiting factor being the abundance of HPV copies and the availability of the target for hybridisation (Robinson et al. 2010). Interpretation of DNA in situ hybridisation shows good inter-observer agreement (Thavaraj et al. 2011; Jordan et al. 2012), and discordance is characterised by weak signals that are often patchy across the tissue section. For negative cases, it is essential that slide-based analyte controls are employed as there are no internal controls to quality assure the adequacy of the staining. Proprietary cell line controls (HistoCyte Laboratories Ltd www.histocyte.com) and tumour xenografts are available (HPV 3 in 1 control, Roche Ventana Medical Systems Ltd). Alternatively, known positive and negative tissue samples can be used as described above. Positive results vary from single punctate signals in the nucleus (Fig 3b), thought to represent single copies of the HPV integrated into the host genome, to cases that have a diffuse pattern of staining located in the nucleus and cytoplasm (Fig. 3c).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree