Chapter 5 T Cell Receptors and MHC Molecules
• The T cell antigen receptor (TCR) is located on the surface of T cells and plays a critical role in the adaptive immune system. Its major function is to recognize antigen and transmit a signal to the interior of the T cell, which generally results in activation of T cell responses.
• TCRs are similar in many ways to immunoglobulin molecules. Both are made up of pairs of subunits (α and β or γ and δ), which are themselves members of the immunoglobulin superfamily, and both recognize a wide variety of antigens via N terminal variable regions. These antigen recognition subunits are associated with the invariant chains of the T cell receptor, the CD3 complex, which perform critical signaling functions.
• The two types of TCR may have distinct functions. In humans and mice, the αβ TCR predominates in most peripheral lymphoid tissues, whereas cells bearing the γδ TCR are enriched at mucosal surfaces.
• Like immunoglobulins, TCRs are encoded by several sets of genes, and a large repertoire of TCR antigen-binding sites is generated by V(D)J recombination during T cell differentiation. Unlike immunoglobulins, TCRs are never secreted and do not undergo class switching or somatic hypermutation.
• Antigen recognition by the αβ TCR requires the antigen to be bound to a specialized antigen-presenting structure known as a major histocompatibility complex (MHC) molecule. Unlike immunoglobulins, TCRs recognize antigen only in the context of a cell–cell interaction.
• Class I and class II MHC molecules bind to peptides derived from different sources. Class I MHC molecules bind to peptides derived from cytosolic (intracellular) proteins, known as endogenous antigens. Class II MHC molecules bind to peptides derived from extracellular proteins that have been brought into the cell by phagocytosis or endocytosis (exogenous antigens).
• Class I and class II MHC present peptide antigens to the TCR in a cell–cell interaction between an antigen-presenting cell (APC) and a T cell.
• In humans the gene loci HLA-A, HLA-B, and HLA-C gene loci encode class I MHC molecules.
• HLA-DP, HLA-DQ, and HLA-DR gene loci encode class II MHC molecules.
• An individual’s MHC haplotype affects susceptibility to disease.
• CD1 is an MHC class I-like molecule that presents lipid antigens.
T cell receptors
As discussed in Chapter 1, the immune system of higher vertebrates can be divided into two components – humoral immunity and cell-mediated immunity.
• targeting them for phagocytosis or complement-mediated lysis;
• neutralizing receptors on the surface of bacteria and viruses; and
TCRs recognize antigen via variable regions generated through V(D)J recombination (see Chapter 3), much like immunoglobulins, but are much more restricted in their antigen recognition capabilities.
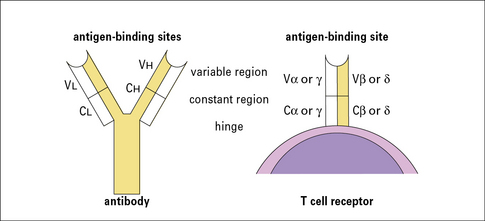
TCRs are similar to immunoglobulin molecules
• resemble immunoglobulins in several significant ways (Fig. 5.1);
• are made up of heterodimers (either α and β or γ and δ subunits), which are disulfide-linked;
• are integral membrane proteins with large extracellular domains and short cytoplasmic tails. The extracellular portions are responsible for antigen recognition and contain variable N terminal regions, like antibodies. Both α and β (or γ and δ) subunits contribute to the antigen-binding sites.
The αβ heterodimer is the antigen recognition unit of the αβ TCR
The αβ TCR is the predominant receptor found in the thymus and peripheral lymphoid organs of mice and humans. It is a disulfide-linked heterodimer of α (40–50 kDa) and β (35–47 kDa) subunits and its structural features have been determined by X-ray crystallography (Fig. 5.2).
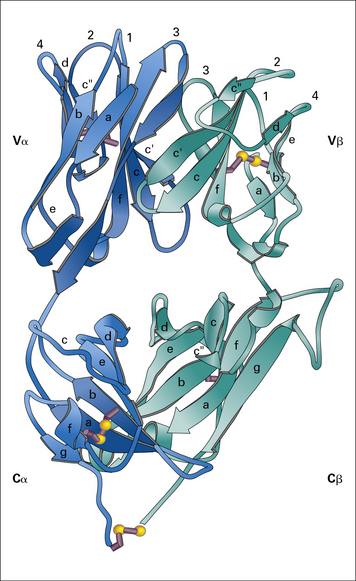
Fig. 5.2 The T cell antigen receptor
(Adapted from Garcia KC, Degano M, Stanfield RL, et al. Science 1996;274:209–219. Copyright AAAS.)
Q. How can receptors that lack intracytoplasmic domains signal to the cell? Give some examples
A. They signal by associating with other membrane molecules that do have intracytoplasmic domains. For example, immunoglobulin associates with Igα and Igβ (see Fig. 3.1), and FcγRI associates with its γ chain dimer (see Fig. 3.17).
The extracellular portions of the α and β chains fold into a structure that resembles the antigen-binding portion (Fab) of an antibody (see Fig. 3.9). Indeed, as in antibodies, the amino acid sequence variability of the TCR resides in the N terminal domains of the α and β (and also the γ and δ) chains.
The regions of greatest variability correspond to immunoglobulin hypervariable regions and are also known as complementarity determining regions (CDRs). They are clustered together to form an antigen-binding site analogous to the corresponding site on antibodies (see Fig. 5.2). Note, however, that:
• the CDR3 loops (which are the most highly variable regions of the TCR, as they are generated by V(D)J recombination) from both the α and β chains lie at the center of the antigen-binding site, and make extensive contacts with antigen.
The disulfide bond that links the α and β chains is in a peptide sequence located between the constant domain of the extracellular portion of the receptor and the transmembrane domain (shown as the C terminal residue in the α and β chain in Fig. 5.2).
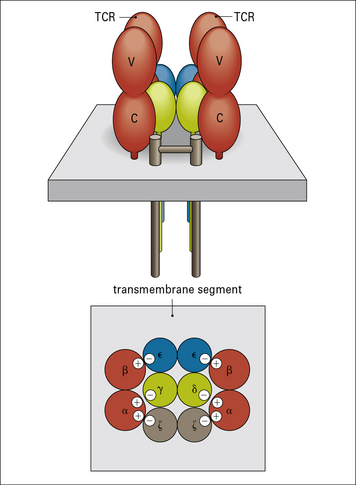
The CD3 complex associates with the antigen-binding αβ or γδ heterodimers to form the complete TCR
The CD3 chains are assembled as heterodimers of γε and δε subunits with a homodimer of ζ chains, giving an overall TCR stoichiometry of (αβ)2, γ, δ, ε2, ζ2. Current data suggest that the TCR complex exists as a dimer (Fig. 5.3).
The negatively charged residues in the transmembrane region of the CD3 chains interact with (and neutralize) the positively charged amino acids in the αβ polypeptides, leading to the formation of a stable TCR complex (see Fig. 5.3).
• a small extracellular domain (nine amino acids);
• a transmembrane domain carrying a negative charge; and
The cytoplasmic portions of ζ and η chains contain ITAMs
The conserved tyrosine residues in the ITAM motifs are targets for phosphorylation by specific protein kinases. When the TCR is bound to its cognate antigen–MHC complex, the ITAM motifs become phosphorylated within minutes in one of the first steps in T cell activation (see Fig. 8.w1![]() ).
).
• are essential for T cell activation, and mutational substitution of the tyrosines in the motif prevents activation;
• play critical roles in B cell activation, and are present in the B cell receptor chains, Igα and Igβ (see Fig. 3.1).
CD3ζ also functions in another signaling pathway, associating with the low-affinity FcγRIIIa receptor (CD16), which is involved in the activation of macrophages and natural killer (NK) cells (see Fig. 3.17).
The γδ TCR structurally resembles the αβ TCR but may function differently
• extracellular V and C domains;
• a transmembrane segment containing positively charged amino acids; and
• in humans and mice, αβ TCRs are present on more than 95% of peripheral blood T cells and on the majority of thymocytes;
• T cells bearing γδ TCRs are relatively rare in spleen, lymph nodes, and peripheral blood but predominate at epithelial surfaces – they are common in skin and in the epithelial linings of the reproductive tract and are especially numerous in the intestine, where they are found as intraepithelial lymphocytes.
Antigen recognition by γδ T cells is unlike that of their αβ counterparts
• γδ T cells can be found in normal numbers in MHC class I and class II-deficient mice;
• their cognate antigens are not necessarily peptides, and do not require classical processing – indeed, some murine γδ T cells have been found to recognize proteins directly, including MHC molecules and viral proteins, in a manner that requires neither antigen processing nor presentation by MHC.
γδ T cells recognize at least two classes of ligand:
• molecules that signal the presence of cellular stress; and
• small organic molecules that serve as signifiers of infection.
γδ T cells have a variety of biological roles
• are essential for primary immune responses to certain viral and bacterial pathogens in mouse models, but in many cases their contribution to the primary response can be substituted by αβ T cells, and they rarely contribute to memory responses;
• interact with a variety of lymphocytes, and have been implicated in stimulating immunoglobulin class switch recombination by B cells in response to T-dependent antigens;
• provide regulatory signals to αβ T cells and have been implicated in shaping immune responses (e.g. γδ cells appear to be involved in downregulating inflammation and in this role may be responding to epithelial cells stressed by inflammatory processes rather than to specific antigens borne by pathogens).
TCR variable region gene diversity is generated by V(D)J recombination
As with antibody genes, a highly diverse repertoire of TCR variable region genes is generated during T cell differentiation by a process of somatic gene rearrangement termed V(D)J recombination (see Chapter 3). Variable (V), joining (J), and sometimes diversity (D) gene segments are joined together to form a completed variable region gene.
The mechanism of V(D)J recombination is the same in both T cells and B cells
The TCR genes are flanked by recombination signal sequences, just like their immunoglobulin cousins (see Fig. 3.24), and the same recombination machinery (the RAG proteins) operates in both B and T cells. Indeed, experiments have shown that TCR Dβ and Jγ genes can rearrange appropriately even if transfected into B cells.
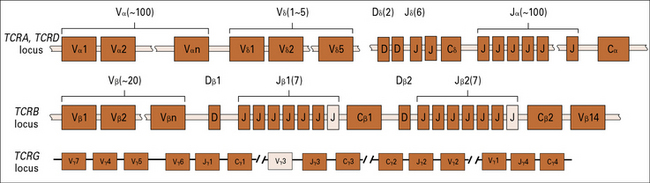
TCRs are encoded by several sets of genes
The general arrangement of the genes encoding the α, β, γ, and δ chains of the TCR is remarkably similar to that of the immunoglobulin heavy chain genes (see Figs 3.21–3.23), suggesting a common origin from a primordial rearranging antigen receptor locus.
Figure 5.w1 illustrates the murine TCR genes, which are similar to those of humans. All four TCR gene families have been strongly conserved across more than 400 million years of evolution of the jawed vertebrates, which suggests a strong selective pressure for the preservation of both αβ and γδ T cell functions.
The TCRB locus includes two sets of D, J, and C genes
The D segments are used in all three reading frames, further contributing to β chain diversity.
The TCRD locus possesses only five Vδ, two Dδ, and six Jδ genes
Q. How may the great number of δ chains be generated when there is only a limited number of V, D, and J gene segments in the TCRD locus?
A. As described in Chapter 3, the two D region segments can be used in all three possible reading frames, and imprecise joining of the VD and DJ junctions produces further diversity.
MHC molecules
Recognition by the αβ TCR requires antigen to be bound to an MHC molecule
As discussed in Chapter 3, antibodies recognize intact proteins, binding either to linear epitopes derived from contiguous amino acids or discontinuous epitopes produced by amino acids that are not near one another in the primary structure but are brought together in the three dimensional structure of the protein. In contrast, T cell receptors only recognize linear epitopes present in the form of short peptides which are generated by degradation of intact proteins within the cell (a process termed antigen processing). This property of T cell receptors is critical to the way the immune system recognizes intracellular pathogens and tumor antigens. Such antigens present a special challenge to the immune system: intracellular antigens are hidden within the cell, and are not available for recognition by antibodies. How can intracellular antigens be recognized by extracellular receptors? The immune system has solved this problem by evolving an elegant means of displaying internal antigens (including those of intracellular pathogens) on the cell surface, allowing their recognition by T cells:
• proteins from within the cell (either produced by the cell or taken into the cell) are digested into short peptide fragments;
• peptides derived from proteins produced within the cell are displayed on the cell surface through binding to specialized antigen-presenting molecules termed MHC class I molecules, which are present on all nucleated cells of the body;
• in a similar fashion, peptides derived from proteins ingested from the extracellular environment by phagocytosis are presented by MHC class II molecules, which are present only on professional antigen presenting cells;
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree