.
b)
The centromere (cen) is designated 10: p10 is the portion of the centromeric band that faces the short arm; q10 is the portion of the centromeric band that faces the long arm.
c)
The numbering of bands starts at the centromere and works toward the ends of the chromosomes. The regions adjacent to the centromere are labeled as 1 in each arm and sequentially (2,3,4…) distally in each chromosomal arm.
d)
Standardized designation of a band requires the: chromosome number; chromosomal arm symbol; the region number and the band number within the region. These are written in order without space or punctuation; the only punctuation (.) is placed after the band if there is/are sub-band(s). For example,
i.
13q21 means chromosome 13, long arm, region 2, band 1 or long arm of chromosome 13, region 2 band 1 (NOT “13 q twenty-one”)
ii.
13q21.3 means chromosome 13, long arm region 2, band 1, sub-band 3 or long arm of chromosome 13, region 2, band 1 sub-band 3 (NOT “13 q twenty-one point three”)
Bands are clearly distinguishable parts of the chromosome by their staining intensity (darker or lighter). A region is “an area of a chromosome lying between two adjacent landmarks” (ISCN, 2013). A landmark is defined as “a consistent and distinct morphologic feature important in identifying chromosomes” (ISCN, 2013).
3)
Basic designation of breakpoints and band composition.
There are two systems of designating structural abnormalities: the short system and the detailed system. Only the short system for designating the breakpoints of structurally abnormal chromosomes will be discussed as this is most relevant to oncology. The breakpoints for the structural alteration are designated in parenthesis immediately following the designation of the type of rearrangement and the chromosome involved (ISCN, 2013; CLSI, 2013).
a)
When both arms of a single chromosome are involved in a two-break rearrangement; the breakpoint in the short “p” arm is always specified before the breakpoint in the long “q” arm.
i.
46,XX,inv(2)(p21q31)
This means that there is a pericentric inversion involving a break in each of the two chromosome arms (“p” and “q”) of the chromosome; in this case the bands at which the breaks and reunion occurred are bands 2p21 and 2q31. When breakage occurs within a single chromosome, the breakpoints are not separated by a semicolon.
b)
When two breaks occur within the same arm (as in the case of EML4-ALK rearrangement), the breakpoint more proximal to the centromere is specified first
i.
46,XX,inv(2)(p21p23)
This means there is a paracentric inversion in which there is breakage in the same arm (“p” in this case) and reunion has occurred at bands 2p21 and 2p23. As noted previously, when both breakpoints are localized to the same chromosome (intrachromosomal rearrangement), they are not separated by a semicolon.
c)
When two chromosomes are involved in a structural rearrangement, “the chromosome having the lowest number is always listed first” if both of the chromosomes are autosomes.
i.
46,XY,t(12;16)(q13;p11.1)
This means that breakage and reunion have occurred at bands 12q13 and 16p11.1. The segments distal to these bands have been exchanged.
d)
If one of the two rearranged chromosomes is a sex chromosome, then the sex chromosome is listed first.
i.
46,X,t(X;18)(p11.1;q11.1)
This means that breakage and reunion have occurred at bands Xp11.1 and 18q11.1. The segments distal to these have bands been rearranged. Note that the correct designation of this rearrangement includes a single structurally normal X chromosome: 46,X,t(X;18)(p11.1;q11.1), only one X is indicated; not 46,XX,t(X;18)(p11.1;q11.1).
4)
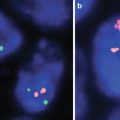
Basic Designation of in situ hybridization (ISH) findings.
There is a difference in nomenclature depending on whether the cell is at the prophase/metaphase versus interphase portion of mitosis. The process of “in situ hybridization” is abbreviated as ISH regardless of whether the method was completed using fluorescent or nonfluorescent methodologies [but fluorescent in situ hybridization (FISH) is a frequently used technique]. Since most of the applications of this technology in solid tumors use interphase in situ hybridization, this will be the focus of this discussion [1, 3–7].
The findings on interphase ISH has a prefix “nuc ish” to indicate that interphase nuclei were evaluated (rather than directly observing metaphase chromosomes). The nomenclature for in situ hybridization also includes the number of probe signals, and, if intrachromosomal, their relative positions. No space or punctuation is used after the “nuc ish” prefix in the description of interphase ISH. Since bands cannot be directly visualized in interphase nuclei, the designation of a chromosomal band is not included in the short nomenclature format used for interphase ISH.
It is important to understand the design and the intended purpose of the probes being evaluated, as this will factor into the interpretation and reporting of the results. There are different types of probes; however, for solid tumors the following are most commonly used:
i)
Locus specific/enumeration probes (e.g., centromeric probes to assess trisomic conditions), which are designed to quantify the number of signals present);
ii)
Fusion probes, which can be single fusion or dual fusion (e.g., for BCR-ABL1), are designed to detect the presence of a rearrangement involving 2 specific loci; or
iii)
Break-apart probes (e.g., for MYC, EWSR1), are designed to detect rearrangements within a gene (This probe is useful in cases where there are multiple translocation partners for a gene with known breakpoints [1, 3, 4, 6].
The following are ISCN recommendations with regard to cytogenetics report for interphase ISH analysis:
A)
Locus specific/enumeration probes:
a.
Number of signals—one probe: immediately following the nuc ish prefix, is a parenthesis having locus designation, a multiplication sign (x) and the number of signals seen:
i.
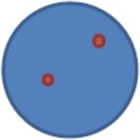
nuc ish(D21S65×2)[200]
This means that there are two copies of locus D21S65 observed in the 200 cells scored
 .
.

ii.
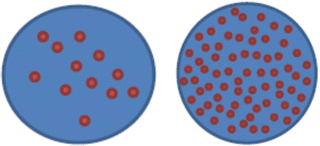
nuc ish(MYCNx12~ > 50)[200]
This means 12 to more than 50 copies of MYCN were observed in the 200 cells scored
 .
.

iii.
nuc ish(MYCN amp)[200]
This means that number of MYCN copies cannot be quantified because it is increased in copy number beyond what can be reliably counted in the 200 nuclei that were scored
 .
.

b.
Number of signals—two or more probes: The probes used in a single hybridization follow one another in a single set of parentheses, separated by a comma sign and a multiplication sign (“×”). If the number of signals is the same for each of the probes this number is indicated outside the parentheses. If the number of signals per probe is different, then each probe has the multiplication sign (“×”) followed by the number of signals, with the findings for each probe being separated by the comma sign [1, 3, 4, 6]. The following are basic examples of the ISCN recommended nomenclature for ERBB2 (commonly known as HER2) ISH:
i.
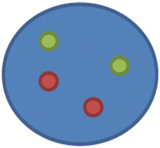
nuc ish(D17Z1,ERBB2)×2[100]
This means that two copies of D17Z1 and ERBB2 were found in the 100 cells scored
 .
.

ii.
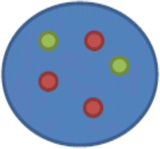
nuc ish(D17Z1x2,ERBB2x3)[60]
This means three copies of ERBB2 were found in the 60 cells scored, but only two copies of the D17Z1 control probe were observed (note that the centromeric probe is listed first because the order of probes is from the tip of the short arm to the tip of the long arm; D17Z1 is localized to the centromere, while ERBB2 is localized to 17q12)
 .
.

iii.
nuc ish(D17Z1x3,ERBB2x9)[100]
This means 9 ERBB2 signals were observed in each of the 100 cells compared to three copies of centromeric 17 probe D17Z1
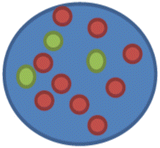 .
.

iv.
nuc ish(D17Z1x2 ~ 3, ERBB2x10 ~ 20)[100/200].
This means 10–20 ERBB2 copies (heterogeneity reflecting the range indicated) were found in 100 (of the total of 200) cells scored compared to 2–3 copies of the centromeric probe for chromosome 17 (D17Z1)
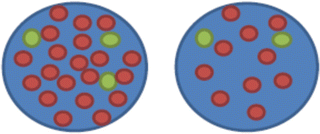 .
.

B)
Fusion probes (single or dual).
If loci on two separate chromosomes are normally separated, fusion of these loci as evidenced by the juxtaposition of the probes, suggests the presence of an abnormality or translocation.
i.
Single fusion probes: Dual color, single fusion probes (usually about 300 kb in size) hybridize to target loci located on each side of a known break-points, with each breakpoint having a different color (e.g., green and red). Typically the breakpoints are located on different chromosomes, such that when there is rearrangement/translocation, the break results in the juxtapositioning of the two colored probes to form a single color (e.g., yellow, from red and green); hence typical abnormal cells will show one red (the structurally normal copy of the first chromosome), one green (the structurally normal copy of the second chromosome) and one yellow signal (a derivative chromosome). This is useful in detecting abnormalities in samples with a high percentage/quantity of translocation. Validation studies are required to quantify the proportion of false positive signals that might occur from the two probes lying too close together (leading to an apparent yellow (fusion) signals) in specimens that lack a rearrangement and to establish cut-off values for interpretation of probe signal patterns [1, 3, 4, 6].
a.
nuc ish(ABL1,BCR)×2[400]
This means there are two copies each of ABL1 and BCR; no fusion in any of the 400 cells scored.
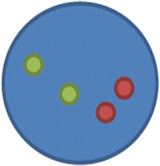

b.
nuc ish(ABL1,BCR)×2(ABL1 con BCRx1)[400]
This means there are two copies each of ABL1 and BCR; however, there is a juxtaposition/fusion (×1) of the ABL1 and BCR loci on a single chromosome.
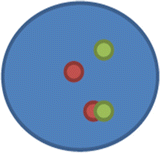

ii.
Dual fusion probes: Dual color, dual fusion probes have lower false positivity rates than single fusion probes. These probes are large probes (300 kb to 1 Mb in size) that overlap the translocation/rearrangement breakpoints that are typically located on two different chromosomes, such that when there is a rearrangement/translocation each probe (one on each chromosome) four fragments are created, with these fragments being fusing to create two separate fusion signals (one on each derivative chromosome). Hence the cell containing a typical balanced translocation will have one red and one green signal (corresponding to the normal chromosomes) and two yellow fusion signals (fusion of the red fragments with the green fragments [1, 3, 4, 6]. It is believed that such fusion patterns are unlikely to occur by chance, reducing the possible false positive signals [3]. Each lab will determine their cut-off values for the probe being used.
a.
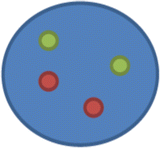
nuc ish(ABL1,BCR)x2[200]
This means that there are two copies each of ABL1 and BCR with no fusion/rearrangement
 .
.

b.
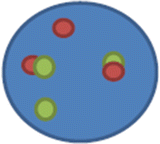
nuc ish(ABL1,BCR)×3(ABL1 con BCRx2)[400].
This means there are three copies each of ABL1 and BCR – there are 3 signals because one red and one green have each split into two at the breakpoints and subsequently formed two derivative (red-green or yellow) chromosomes; the rearrangement caused juxtapositioning/fusion (×2) of the ABL1 and BCR loci on each of the two derivative chromosomes.
ABL1 (c-abl oncogene 1, non-receptor tyrosine kinase); BCR (breakpoint cluster region).

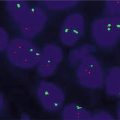
c)
Break–apart probes: If the loci are normally juxtaposed and appear fused because of the closeness of the loci on the same chromosome, separation of these loci using break-apart probes suggests rearrangement. Break-apart probes have two differentially colored probes that hybridize to targets on opposite side of a translocation/rearrangement breakpoint in one gene (on one chromosome) as opposed to fusion probes which hybridize to two different genes (on two different chromosomes). In cells without a translocation/rearrangement, the two probes are so close together that the colors appear as a single fusion signal (e.g., red and green appear as one yellow signal). However, in cells with a translocation/rearrangement, there is a separation at the break-point, leading to clear separation of the probes or signals (green and red). Hence one set of probes is split into green and red (rearranged), while the other set is still close (normal). In order to minimize false positive interpretation, the signals should be at least one to two signals apart, to be considered break-apart or positive.
i.
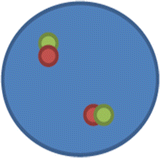
nuc ish(SS18x2)[200].
This means that there are two SS18 signals in the 200 cells scored, with no separation of the signals/probe; hence this is negative for a rearrangement (inversion, translocation) of the SS18 (synovial sarcoma translocation, chromosome 18)
 .
.

ii.
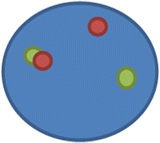
nuc ish(SS18x2)(5′SS18 sep 3′SS18x1)[175/200]
This means that there are two SS18 signals, but one has separated into the 5′ probe and the 3′ probe; hence in 175 of the 200 nuclei scored, the pattern is positive for a rearrangement of the SS18 locus (synovial sarcoma translocation, chromosome 18)
 .
.

Reference
1.
ISCN (2013): An international system for human cytogenetic nomenclature. Basel, Switzerland: S. Karger; 2013.
2.
Simons A, Shaffer LG, Hastings RJ. Cytogenetic nomenclature: changes in the ISCN 2013 compared to the 2009 edition. Cytogenet Genome Res. 2013.
3.
CLSI. Fluorescence in situ hybridization methods for clinical laboratories; approved guideline – second edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2013.
4.
Cook JR. Paraffin section interphase fluorescence in situ hybridization in the diagnosis and classification of non-Hodgkin lymphomas. Diagn Mol Pathol B. 2004;13(4):197–206.
5.
Raimondi SC. Fluorescence in situ hybridization: molecular probes for diagnosis of pediatric neoplastic diseases. Cancer Invest. 2000;18(2):135-47.
6.
Tibiletti MG. Interphase FISH as a new tool in tumor pathology. Cytogenet Genome Res. 2007;118(2-4):229–36.
7.
Waters JJ, Barlow AL, Gould CP. Demystified … FISH. Mol Pathol. 1998;51(2):62–70.
In solid tumors the most common cytogenetic testing is FISH and the majority of the specimens are formalin-fixed paraffin-embedded (FFPE) tissues, hence interphase FISH is used. Generally, the basic FISH techniques include: prehybridization (deparaffinization and aggressive proteolytic treatments to remove unwanted proteins/autofluorescence and allow the probe to interact with the target); denaturation of the target and the probe DNAs using formamide at a high temperature (a variety of probes may be used); and signal detection using a different combination of fluorochrome conjugated antibodies [55–58]. Different types of probes are available for FISH testing [55–59]. The ones commonly used for solid tumors are highlighted in Box 3.1
The results of FISH and other cytogenetic studies should be reported in a standardized format. The use of most updated nomenclature is strongly recommended and required for diagnostic cytogenetic reports, cytogenetic publications, proficiency testing and for laboratory accreditation [7, 19, 20]. Box 3.1 highlights some examples of standardized cytogenetic nomenclature as recommended by ISCN [19, 20].
Standardized Nomenclature: Gene Name [22, 46, 60–63]
The HGNC has approved standardized gene names and unique symbols for genes. The gene name gives information on the character/function of the genes. The gene names, however, may be long and may be impractical for daily or practical usage, hence the need for unique symbols for gene names. For example, it is difficult to use the approved gene name “phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha,” however, the approved unique symbol for that gene “PIK3CA” is relatively easy to use. The HGNC approved gene name which attempts to capture the functions [64] of the gene (“name of function”) is likely to become the legacy name; this should be distinguished from names of the gene products/name of molecule or proteins (which uses the same gene symbols). The criteria or rationale used for giving gene names and symbols are indicated on the HGNC website [61]. Given the ingrained nature of the use of colloquial names, there may be reluctance to transition to or use the standard nomenclature recommended by HGNC and some have argued that changing such colloquial names (aliases) to the standard nomenclature may lead to confusion. To minimize confusion during the transition from the use of colloquial names to the use of standard nomenclature, the common names/aliases may be put in parentheses as highlighted in the last column in Table 3.1. HGNC recommends the use of italics for gene symbols (in upper case letters), while proteins have the same symbols (upper case letters), e.g., KRAS (gene), KRAS (protein). Italicizing gene symbols applies to mRNA, genomic DNA and cDNA, with relevant prefix.; it also applies to pseudogenes and non-coding RNA. It must be emphasized however, that italicizing gene symbols is not universal and some (including some journals) may refer to gene symbols in a non-italicized format.
Table 3.1
Standard nomenclature and aliases of common genes
Solid tumors | |||||
|---|---|---|---|---|---|
Commonly used gene symbols and names | Other aliases | HGNC approved name | HGNC approved gene symbol (standard nomenclature) | Chromosomal location/why important | HGNC recommended protein symbol (Note: protein symbols are the Same as Gene but not italicized) |
ALK | CD246, NBLST3 | Anaplastic lymphoma receptor tyrosine kinase | ALK | 2p23 May be confused with ALK1 (if improperly used) | ALK |
ALK1 | ALK-1, HHT, HHT2, ORW2, ACVRLK1, SK3, TSR-1 | Activin A receptor type II-like 1 | ACVRL1 | 12q13.13 May be confused with ALK. | |
AKT3 | PKBG, PRKBG, “protein kinase B, gamma,” RAC-gamma, MPPH, RAC-PK-gamma, STK-2 | v-akt murine thymoma viral oncogene homolog 3 | AKT3 | 1q44 | AKT3 |
BRAF | BRAF1, B-RAF1, NS7, RAFB1 | v-raf murine sarcoma viral oncogene homolog B | BRAF | 7q34 | BRAF |
BRCA1 | BRCC1, PPP1R53, RNF53, BROVCA1, IRIS, PNCA4, PSCP | Breast cancer 1, early onset | BRCA1 | 17q21.31 | BRCA1 |
BRCA2 | BRCC2, FAD, FAD1, BROVCA2, FACD, FANCD1, GLM3, PNCA2 | Breast cancer 2, early onset (previous symbols and names: FACD, FANCD, FANCD1, “Fanconi anemia, complementation group D1”) | BRCA2 | 13q12-q13 | BRCA2 |
Calponin | Sm-Calp, SMCC, HEL-S-14, | Calponin 1, basic, smooth muscle | CNN1 | 19p13.2-p13.1 | CNN1 |
Calretinin | CAL2, “calretinin” | Calbindin 2 | CALB2 | 16q22.2 | CALB2 |
CDX2 | Caudal type homeobox 2 (previous symbols and names: “caudal type homeo box transcription factor 2,” CDX3) | CDX2 | 13q12.2 | CDX2 | |
CHOP | C/EBP zeta, CHOP, CHOP10, CHOP-10, GADD153 | DNA-damage-inducible transcript 3 | DDIT3 | 12q13.1-q13.2 | DDIT3 (aka CHOP) |
Chromogranin | “Pancreastatin,” “parastatin,” “vasostatin,” CGA | Chromogranin A (parathyroid secretory protein 1) | CHGA | 14q32 | CHGA |
Chromogranin | “Secretogranin B” SCG1 | Chromogranin B (secretogranin 1) | CHGB | 20p12.3 | CHGB |
c-MET | Hepatocyte growth factor receptor, (HGFR), RCCP2 | Met proto-oncogene | MET | 7q31 | MET (aka HGFR, RCCP2) |
COL1A1 | OI4 | Collagen, type I, alpha 1 | COL1A1 | 17q21.33 | COL1A1 |
Desmin | CMD1I, CSM1, CSM2, “intermediate filament protein,” LGMD2R | Desmin | DES | 2q35 | DES |
DPC4 | JIP, MADH4, MYHRS | SMAD family member 4 | SMAD4 | 18q21.1 | SMAD4 (aka DPC4) |
E-cadherin | CD324, “E-Cadherin,” uvomorulin | Cadherin 1, type 1, E-cadherin (epithelial) (previous symbols and names: UVO) | CDH1 | 16q22.1 | CDH1 |
EGFR | ERBB, ERBB1 | Epidermal growth factor receptor | EGFR | 7p12 | EGFR |
ER | ESR, ESRA, ESTRR, Era, NR3A1, RP1-130E4.1, | Estrogen receptor 1 | ESR1 | 6q24-q27 ER can mean “endoplasmic reticulum,” “emergency room.” There is also ESR2 related to ESR1. There are 28 “ER” associated genes in genenames.org, but only one “ESR1” | ESR1 (aka ER) |
ETV6 | TEL, TEL oncogene, TEL/ABL | ETS variant 6 | ETV6 | 12p13 | ETV6 |
EWS | EWSR1, AC002059.7, bK984G1.4 | EWS RNA-binding protein 1 (previous name: Ewing sarcoma breakpoint region 1) | EWSR1 | 22q12.2 | EWSR1 (aka EWS) |
EWSR1 | EWS, AC002059.7, bK984G1.4 | EWS RNA-binding protein 1 (previous name: Ewing sarcoma breakpoint region 1) | EWSR1 | 22q12.2 | EWSR1 (aka EWS) |
HER2 | CD340, HER-2, HER-2/neu, NEU, MLN 19, NGL, TKR1, neuro/glioblastoma derived oncogene homolog | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | ERBB2 | 17q11.2-q12 Although HER2 is widely known, the use of the standard nomenclature removes ambiguity and makes for uniformity of literature review | ERBB2 (aka HER2, HER-2) |
KRAS | KRAS1, KRAS2, C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, NS, NS3, RASK2 | Kirsten rat sarcoma viral oncogene homolog | KRAS | 12p12.1 | KRAS |
LKB1 | PJS, hLKB1, polarization-related protein LKB1, | Serine/threonine kinase 11 | STK11 | 19p13.3 | STK11 (aka LKB1) |
MYF4 | bHLHc3, myf-4 | Myogenin (myogenic factor 4) (previous symbols and names: MYF4) | MYOG | 1q31-q41 | MYOG (aka MYF4) |
MYOD1 | bHLHc1, “myoblast determination protein 1,” MYOD, PUM, MYF3 | Myogenic differentiation 1 (previous symbols and names: MYF3, “myogenic factor 3”) | MYOD1 | 11p15 | MYOD1 |
NTRK3 | TRKC, gp145 | Neurotrophic tyrosine kinase, receptor, type 3 | NTRK3 | 15q24-q25 | NTRK |
NRAS | RP5-1000E10.2, ALPS4, N-ras, NRAS1, NS6, HRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | NRAS | 1p13.2 | NRAS |
NUT | DKFZp434O192, FAM22H, “nuclear protein in testis,” C15orf55 | NUT midline carcinoma, family member 1 | NUTM1 | 15q14 | NUTM1 (aka NUT) |
PAX3 | HUP2, CDHS, WS1, WS3 | Paired box 3 | PAX3 | 2q36.1 | PAX3 |
PAX7 | HUP1, PAX-7, RP23-334I5.3 | Paired box 7 | PAX7 | 1p36.13 | PAX7 |
PAX8 | Paired box 8 | PAX8 | 2q13 | PAX8 | |
PDGFB | “becaplermin,” “oncogene SIS,” SSV, LL22NC03-10C3.2, IBGC5, PDGF-2, PDGF2, SIS, c-sis | Platelet-derived growth factor beta polypeptide | PDGFB | 22q13.1 | PDGFB |
PDGFRA | CD140a, PDGFR2, PDGFR-2, RHEPDGFRA | Platelet-derived growth factor receptor, alpha polypeptide | PDGFRA | 4q10 | PDGFRA |
PIK3CA | PI3K, CLOVE, CWS5, MCAP, MCM, MCMTC, p110-alpha | Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha | PIK3CA | 3q26.3 | PIK3CA |
PIK3R1 | GRB1, p85, p85-ALPHA, AGM7 | Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | PIK3R1 | 5q13.1 | PIK3R1 |
PPARγ1 | CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | Peroxisome proliferator-activated receptor gamma | PPARG | 3p25 | PPARG |
PR | NR3C3 | Progesterone receptor | PGR | 11q22-q23 “PR” can mean prolactin receptor. There are 25 “PR” associated genes in genenames.org, but only one “PGR” | PGR (aka PR) |
RET | CDHF12, CDHR16, HSCR1, MEN2A, MEN2B, MTC1, PTC, RET-ELE1, RET51 | RET proto-oncogene | RET | 10q11.2 | RET |
SMAD4 | JIP, MADH4, MYHRS, DPC4 | SMAD family member 4 | SMAD4 | 18q21.1 | SMAD4 (aka DPC4) |
SSX1 | RP11-552E4.1, CT5.1, SSRC | Synovial sarcoma, X breakpoint 1 | SSX1 | Xp11.23 | SSX1 |
SSX2 | RP11-552J9.2, CT5.2, CT5.2A, HD21, HOM-MEL-40, SSX | Synovial sarcoma, X breakpoint 2 | SSX2 | Xp11.22 | SSX2 |
STK11 | LKB1, PJS, hLKB1, polarization-related protein LKB1, MGC119055, MGC15364, MGC3884 | Serine/threonine kinase 11 | STK11 | 19p13.3 | STK11 (aka LKB1) |
SYT | SSXT | Synovial sarcoma translocation, chromosome 18 | SS18 | 18q11.2 | SS18 (aka SYT/SSXT) |
TFE3 | RCCP2, RCCX1, TFEA, bHLHe33 | Transcription factor binding to IGHM enhancer 3 | TFE3 | Xp11.22 | TFE3 |
TTF1 | TTF-1, TTF-I | Transcription termination factor, RNA polymerase I NOTE: This is unrelated to thyroid transcription factor. See NKX2 – 1 for the IHC used for lung cancer diagnosis | TTF1 | 9q34.3 This is unrelated to Thyroid transcription factor (NK2 homeobox 1 gene) used lung adenocarcinoma diagnosis. Using TTF-1 for that gene and protein product is a misnomer that could lead to confusion | TTF1 |
TTF-1 | TTF-1, TTF1, BCH, BHC, NK-2, NKX2.1, NKX2A, TEBP, TITF1, | NK2 homeobox 1 (previous symbols and names: BCH, “benign chorea,” NKX2A, “thyroid transcription factor 1,” TITF1) | NKX2-1 | 14q13.3 | NKX2-1 (aka TTF-1) |
BCL1 | “B-cell CLL/lymphoma 1,” “G1/S-specific cyclin D1,” “parathyroid adenomatosis 1,” U21B31, D11S287E, PRAD1, | Cyclin D1 (previous symbols and names: BCL1, “cyclin D1 (PRAD1: parathyroid adenomatosis 1),” D11S287E, PRAD1) | CCND1 | 11q13 | CCND1 |
BCL2 | Bcl-2, PPP1R50, “protein phosphatase 1, regulatory subunit 50” | B-cell CLL/lymphoma 2 | BCL2 | 18q21.3 | BCL2 |
BCL6 | BCL5, BCL6A, LAZ3, ZBTB27, ZNF51 | B-cell CLL/lymphoma 6 (previous symbols and names: “zinc finger protein 51,” ZNF51) | BCL6 | 3q27 | BCL6 |
BCR | Breakpoint cluster region (previous symbols and names: BCR1, D22S11) | BCR | 22q11 | BCR | |
c-KIT | C-Kit, CD117, SCFR, PBT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (previous outdated name: BT, “piebald trait” | KIT | 4q12 | KIT (aka C-Kit) |
c-MYC | MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog | MYC | 8q24 | MYC |
FGFR1 | FGFR, CD331, CEK, FLG, H2, H3, H4, H5, N-SAM, “Pfeiffer syndrome,” BFGFR, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | Fibroblast growth factor receptor 1 | FGFR1 | 8p12 | FGFR1 |
FGFR3 | HBGF-3, “INT-2 proto-oncogene protein,” “murine mammary tumor virus integration site 2, mouse,” “oncogene INT2,” “V-INT2 murine mammary tumor virus integration site oncogene homolog” | Fibroblast growth factor 3 (previous symbols and names: “fibroblast growth factor 3 (murine mammary tumor virus integration site (v-int-2) oncogene homolog),” INT2) | FGF3 | 11q13 | FGF3 |
IGH | IGD1, IGH.1@, IGH@, IGHD@, IGHDY1, IGHJ, IGHJ@, IGHV, IGHV@ | Immunoglobulin heavy locus | IGH | 14q32.33 | IGH |
MALT1 | IMD12, MLT, MLT1, “paracaspase” | Mucosa associated lymphoid tissue lymphoma translocation gene 1 (previous symbols and names: MLT) | MALT1 | 18q21 | MALT1 |
MLL | hCG_1732268, ALL-1, CXXC7, HRX, HTRX1, MLL, MLL/GAS7, MLL1, MLL1A, TET1-MLL, TRX1, WDSTS | Lysine (K)-specific methyltransferase 2A (previous/outdated name: MLL, “myeloid/lymphoid or mixed-lineage leukemia (trithorax (Drosophila) homolog),” “myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)” | KMT2A | 11q23 | KMT2A (aka MLL) |
MMSET | MMSET, NSD2, RP11-262P20.3, REIIBP, TRX5, WHS | Wolf-Hirschhorn syndrome candidate 1 (previous symbols and names: Multiple myeloma SET domain-containing protein Short name = MMSET) | WHSC1 | 4p16.3 | WHSC1 |
MYH11 | SMHC, SMMHC, AAT4, FAA4, | Myosin, heavy chain 11, smooth muscle | MYH11 | 16p13.11 ? Similarity and confusion with SMMHC used for myoepithelial cells in breast pathology | MYH11 |
HGNC also indicated that “in addition the names of genes coding for enzymes should be based on those recommended by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology”[32, 33]. Furthermore, “names of genes encoding plasma proteins, hemoglobins and specialized proteins” are based on standard names (as recommended by HGNC) as well as those recommended by their respective committees [46, 61], this may not be unrelated to the maintenance of the “legacy numbering schemata” of these other committees [65, 66]. Nevertheless, adopting a uniform nomenclature system will allow for uniform or easier update, if necessary in the future. The importance of sticking to a uniform nomenclature/symbol can be emphasized by this argument: In a vast majority of settings in molecular/medical science, we are examining proteins as just one isoform. For example, at the present time, the ESR1 protein (estrogen receptor 1) is adequate; however, the same protein is also called ER alpha or just ER. In the future, however, given the advances in medical/molecular science, it is conceivable that other isoforms of peptides or proteins may be discovered. The discovery of other isoforms will necessitate updating the current nomenclature to accommodate the other isoforms that may be discovered to be clinically useful; these new isoforms will need to be distinguished from the currently tested ESR1 isoform. Having different names for the same entity will complicate nomenclature of these isoforms, if and when needed. Using or having different names for estrogen receptors (ESR1 vs. ER alpha vs. ER) may complicate naming these isoforms. ER-alpha’s isoforms may be named in ways which will likely be completely different from the naming of “ESR1” isoforms. This possible confusion can be eliminated or minimized by sticking to a uniform nomenclature system—ESR1 can be used as the stem symbol for the other isoforms, if this symbol is uniformly used for estrogen receptor 1.
Standardized Nomenclature: Description of Sequence Variant [39, 67–70]
In order to understand standard nomenclature of sequence variant, a review of basic molecular pathology including terminologies in Chaps. 1 and 2 of this text is recommended. Understanding the basic terminologies will enhance understanding of the basis for sequence variant nomenclature.
It is important to know that gene nomenclature continues to evolve with new technology [71]. Standard nomenclature of description of sequence variant is usually given at the DNA level in relation to a reference sequence—either genomic or a coding DNA reference sequence [49, 65, 67, 70, 72–76]. When changes at the RNA or protein are given, the changes must be described at the DNA level first, with RNA/protein changes given in parentheses, for example, BRAF mutation will be described as: NM_004333.4: c.1799T > A (p.Val600Glu); including the protein change is useful but not mandatory. Some practical recommendations are described in Box 3.2.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree