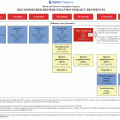
Study
GWAS vs. candidate gene
Study design
Replication
Sample size
Results
Therapy-related leukemia-associated with exposure to alkylating agents and topoisomerase II inhibitors
Genome-wide association studies
Knight et al. 2009 [90]
GWAS
Case–control (healthy controls)
Yes
Discovery set: 80 cases; 150 controls
Among patients with acquired abnormalities of chromosomes 5 or 7; 3 SNPs (rs1394384 [OR = 0.3, 95 % CI, 0.2–0.6], rs1381392 [OR = 2.1, 95 % CI, 1.3–3.4], and rs1199098 [OR = 0.5, 95 % CI, 0.3–0.8]) were associated with t-MDS/AML
Replication set: 70 cases; 95 controls
Candidate gene approach
Ellis et al. 2008 [89]
Candidate gene (2 common functional p53-pathway variants, the MDM2 SNP309 and the TP53 codon 72
Case–control (healthy controls)
Yes
Discovery set: 80 cases
Neither polymorphism alone influenced the risk of t-MDS/AML; however an interactive effect was detected such that MDM2 TT TP53 Arg/Arg double homozygotes, and individuals carrying both a MDM2 G allele and a TP53 Pro allele, were at increased risk of t-MDS/AML (OR = 2.0, 95 % CI, 1.2–3.5, Pinteraction = 0.009)
Replication set: 91 cases
Allan et al. 2001 [61]
Candidate gene approach
Case–control
No
89 cases; 420 patients with de novo AML; 1,022 healthy controls
Individuals with at least one GSTP1 codon 105 Val allele were over-represented in t-AML cases compared with de novo AML cases (OR = 1.8, 95 % CI, 1.1–2.9). Compared with de novo AML, the GSTP1 codon 105 allele occurred more often among t-AML patients with prior exposure to chemotherapy (OR = 2.7, 95 % CI, 1.4–5.1), particularly among those with prior exposure to known GSTP1 substrates (OR = 4.3, 95 % CI, 1.4–13.2)
Polymorphisms in GSTM1, GSTT1, GSTP1
Worrillow et al. 2003 [67]
Candidate gene
Case–control
No
91 cases; 420 patients with de novo AML, 837 healthy controls
The variant (C) hMSH2 allele was significantly overrepresented in t-AML cases that had previously been treated with O6-guanine alkylating agents, including cyclophosphamide and procarbazine, compared with controls (OR = 4.0, 95 % CI 1.4–11.4); 38 % of the patients were MSI positive
hMSH2 –6exon 13 polymorphism
Verification performed by direct sequencing
Evaluation of MSI
Worrillow et al. 2008 [98]
Candidate gene
Case–control
No
133 cases
Carrier frequency of MLH1-93 variant was higher in patients who developed t-AML after alkylating agents exposure for HL, compared to patients without alkylating agent exposure. The MLH1 -93 variant allele was also over-represented in t-AML cases when compared to de novo AML cases and was associated with increased risk of t-AML (OR = 5.31, 95 % CI, 1.40–20.15) among patients exposed to alkylating agents
Polymorphism of MLH1 (position −93, rs1800734)
420 patients with de novo AML, 242 patients with primary HL, 1,177 healthy controls
Seedhouse et al. 2002 [86]
Candidate gene
Case–control
No
34 cases; 134 patients with de novo AML; 178 healthy controls
Presence of at least one XRCC1 399Gln allele indicated a protective effect for the allele in controls compared with patients with t-AML (OR = 0.4, 95 % CI, 0.2–0.9)
Polymorphisms in XRCC1, XRCC3, XPD, NQO1
Jawad et al. 2006 [74]
Candidate gene
Case–control
No
42 cases; 166 patients with de novo AML; 189 healthy controls
Presence of the variant HLX1 allele significantly increased the risk of t-AML (OR = 3.4, 95 % CI, 1.7–6.8). Polymorphism in RAD51 (135G/C-5′ UTR) also increased the risk of t-AML. Combined analysis revealed, a synergistic 9.5-fold increase (95 % CI, 2.2–40.6) in risk for t-AML
C/T-3′ untranslated region (UTR) polymorphism in HLX; Polymorphism in RAD51 (135G/C-5′ UTR)
Subsequent solid malignancies–associated with exposure to radiation therapy
Genome-wide association studies
Best et al. 2011 [91]
GWAS
Case–control (controls: cancer survivors with no SMNs)
Yes
Discovery set: 100 cases; 89 controls
Two variants at chromosome 6q21 (rs4946728 [OR = 11.4, 95 % CI, 3.2–40.3]; rs1040411 [OR = 6.6, 95 % CI, 3.2–13.5]) were associated with SMNs in childhood Hodgkin lymphoma (HL) survivors, but not in adult-onset HL survivors
Replication set: 62 cases; 71 controls
Candidate gene approach
Mertens et al. 2004 [99
]
Candidate gene
Cohort study
No
650 patients with HL (178 with subsequent malignancy)
Individuals lacking GSTM1 were at increased risk of any subsequent malignancy (OR = 1.5, 95 % CI, 1.0–2.3). A non-significant increased risk for thyroid cancer was observed in individuals lacking either GSTM1 (OR, 2.9, 95 % CI, 0.8–10.9) or GSTT1 (OR, 3.7, 95 % CI, 0.6–23.5). Individuals with the genotype of the Arginine/glutamine polymorphism at codon 399 in the XRCC1 gene (R399)r showed a nonsignificant increased risk of breast cancer (OR, 1.4, 95 % CI, 0.7–2.7)
Polymorphisms in GSTM1, GSTT1, XRCC1
Using a case–control study design, Ellis et al. examined the association between t-MDS/AML and 2 common functional p53-pathway variants—the MDM2 SNP309 and the TP53 codon 72 polymorphism [89]. While neither polymorphism demonstrated a significant association individually, an interactive effect was detected such that individuals carrying both a MDM2 G allele and a TP53 Pro allele were at increased risk of chemotherapy-related t-MDS/AML.
Knight et al. used a case–control study design, to conduct a Genome-Wide Association Study (GWAS) in patients with t-MDS/AML [90]. The discovery set included 80 cases and 150 healthy controls; relevant findings were replicated in an independent set of 70 cases and 95 healthy controls. The investigators identified three SNPs (rs1394384 [OR = 0.3, 95 % CI, 0.2–0.6], rs1381392 [OR = 2.1, 95 % CI, 1.3–3.4], and rs1199098 [OR = 0.5, 95 % CI, 0.3–0.8]) to be associated with t-MDS/AML with chromosome 5/7 abnormalities. rs1394384 is intronic to ACCN1, a gene encoding an amiloride-sensitive cation channel that is a member of the degenerin/ epithelial sodium channel; rs1199098 is in LD with IPMK, which encodes a multikinase that positively regulates the prosurvival AKT kinase and may modulate Wnt/beta-catenin signaling; rs1381392 is not near any known genes, miRNAs, or regulatory elements, although it lies in a region recurrently deleted in lung cancer. Although the investigators were able to confirm findings in an independent replication cohort, utilization of a non-cancer healthy control group raises concerns about the case–control differences being generated by genetic susceptibility to the primary cancer vs. t-MDS/AML.
Best et al. performed a GWAS to identify variants associated with radiation-related solid malignancies in HL survivors [91]. They identified two variants at chromosome 6q21 associated with SMNs. The variants comprise a risk locus associated with decreased basal expression of PRDM1 and impaired induction of the PRDM1 protein after radiation exposure. These data suggest new gene-exposure interaction that may implicate PRDM1 in the etiology of radiation therapy-induced second malignancies.
Studies such as those described above are being increasingly described. Large populations, with well-validated cases, appropriate controls, and good quality biospecimens are needed to understand the pathogenesis of SMNs. Furthermore, it is critical to replicate the findings in clinically independent cohorts, and also examine the functional relevance of the genes implicated in the development of SMNs. Nonetheless, understanding the pathogenesis of SMN will facilitate focused medical follow-up care and surveillance of the ever-growing population of childhood cancer survivors.
14.3 Screening
Several groups have developed recommendations for cancer surveillance on the basis of therapeutic exposures, recognizing that these cancers are a leading cause of non-relapse late mortality [92] and serious morbidity [93]. The underlying premise for these surveillance recommendations is that almost half of the SMNs can be detected at an early stage by periodic surveillance [1, 3, 94, 95]. In general, because survival rates are strongly associated with the stage of disease at diagnosis, early initiation of SMN surveillance is recommended. This early surveillance is even more necessary because options become limited for the treatment of SMNs because of prior therapeutic exposures. The COG LTFU Guidelines [96] recommend monitoring for t-MDS/AML with annual complete blood count for 10 years after exposure to alkylating agents or topoisomerase II inhibitors. Most other SMNs are associated with radiation exposure, present as solid tumors and are site-specific. Screening recommendations include careful annual physical examination of the skin and underlying tissues in the radiation field. For example, young women treated for a childhood cancer with a moderate-to-high dose of chest radiation have a greatly increased risk of breast cancer, similar in magnitude to that of carriers of the BRCA mutation [97]. Mammography, the most widely accepted screening tool for breast cancer in the general population, may not be the ideal screening tool by itself, for radiation-related breast cancers occurring in relatively young women with dense breasts, hence the American Cancer Society recommends adjunct screening with MRI. Thus, the COG LTFU recommendations for females who received radiation with potential impact to the breast (i.e., radiation doses of 20 Gy or higher to the mantle, mediastinal, whole lung, and axillary fields) include monthly breast self-examination beginning at puberty; annual clinical breast examinations beginning at puberty until age 25 years; and a clinical breast examination every 6 months, with annual mammograms and MRIs beginning 8 years after radiation or at age 25 (whichever occurs later). Screening of those at risk for early-onset colorectal cancer (i.e., radiation doses of 30 Gy or higher to the abdomen, pelvis, or spine) should include colonoscopy every 5 years beginning at age 35 years or 10 years following radiation (whichever occurs last).
14.4 Conclusion
Our knowledge is constrained by the follow-up of childhood cancer survivors that does not extend beyond three decades. It is only with the accrual of adequate numbers of survivors in their 4th and 5th decade after diagnosis of primary cancer will we learn how the normal ageing processes and the natural increase of cancer in the general population influences the development of SMNs in the ageing cohort of childhood cancer survivors.
More research is needed to understand the pathogenesis of treatment-related cancers, and to characterize those individuals at highest risk. This information needs to be used to develop cancer-risk prediction models to estimate individual risk, and help discussions between the healthcare provider and the survivor about surveillance for SMNs. There is a critical need to promote and optimize screening for SMNs, and develop behavioral and pharmacologic intervention trials to stop or reverse the process of progression of premalignant lesions into overt malignant diseases. Finally, there is an equally critical need to use this information to modify upfront therapy, while balancing cure with long-term morbidity.
References
1.
2.
Reulen, R. C., Frobisher, C., Winter, D. L., et al. (2011). Long-term risks of subsequent primary neoplasms among survivors of childhood cancer. JAMA, 2011(305), 2311–2319.CrossRef
3.
Friedman, D. L., Whitton, J., Leisenring, W., et al. (2010). Subsequent neoplasms in 5-year survivors of childhood cancer: The Childhood Cancer Survivor Study. Journal of the National Cancer Institute, 102, 1083–1095.PubMedCentralPubMedCrossRef
4.
Armstrong, G. T., Liu, W., Leisenring, W., et al. (2011). Occurrence of subsequent neoplasms in long-term survivors of childhood cancer: A report from the Childhood Cancer Survivor Study. Journal of Clinical Oncology, 29, 3056–3064.PubMedCentralPubMedCrossRef
5.
6.
7.
8.
9.
10.
Swerdlow, A. J., Barber, J. A., & Hudson, G. V. (2000). Risk of second malignancy after Hodgkin’s disease in a collaborative British cohort: The relation to age at treatment. Journal of Clinical Oncology, 18, 498–500.PubMed
11.
Bhatia, S., Krailo, M. D., Chen, Z., et al. (2007). Therapy-related myelodysplasia and acute myeloid leukemia after Ewing sarcoma and primitive neuroectodermal tumor of bone: A report from the Children’s Oncology Group. Blood, 109, 46–51.PubMedCentralPubMedCrossRef
12.
13.
14.
15.
Travis, L. B., Holowaty, E. J., Bergfeldt, K., et al. (1999). Risk of leukemia after platinum-based chemotherapy for ovarian cancer. The New England Journal of Medicine, 340, 351–357.PubMedCrossRef
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree