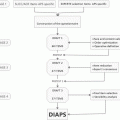
Mouse model
Comments
Features of APS
Production of aPL
MRL/Mp-lpr/lpr (MRL/lpr) [2]
–
Yes
Yes
MRL-lpr/lpr (MRL/+) [2]
–
Yes
Yes
NZB [3]
–
Yes
Yes
BXSB-Yaa [4]
Features are of decreased frequency and intensity than in W/B F1 male progeny
Yes, males
Yes, males
NZW (female) × BXSB (male) F1 (W/B F1) [5]
–
Yes, males
Yes, both genders
C57BL/6 J [6]
Augmented by estrogen supplementation
No
Yes
AKR/J [7]
Autoimmune diabetes model
No
Yes, transient
NOD [8]
Autoimmune diabetes model
No
Yes
Another study used an annexin A5 knockout mouse model (Anxa5-KO) to evaluate its role in pregnancy-related morbidity [11]. The litter size was significantly reduced by deficient maternal annexin A5 production, with evidence of placental thrombi formation and fetal growth restriction; these findings were ameliorated with treatment with heparin. These results support the hypothesis that the maternal supply of annexin A5 to the circulation is necessary for maintaining a fully intact pregnancy. An additional study examined the association of prothrombotic factor V Leiden (FVL) on APS manifestations [12]. Evaluating a mouse model of central nervous system manifestations of APS with a knock-in transgene for FVL, an increase in aPL levels and a number of behavioral/cognitive dysfunction and neurodegenerative changes associated with these autoantibodies were noted in the FVL APS mice. These effects were linked to gene dosage, and were thus significantly more pronounced in homozygous than in heterozygous mice, supporting the synergistic impact of other thrombotic risk factors in the manifestations of APS.
Antiphospholipid Syndrome Family and Population Studies
Clinical Phenotype Evaluations
Family and population studies may describe clinical phenotypes, including laboratory values, or may delve further into genotypes, the latter more systematically assessing inherited risk for APS. In exploring clinical phenotypes, family history identifies individuals with clinical manifestations associated with the syndrome [13]. An inherent limitation to this method is that by collecting retrospective clinical data without prospective confirmation, one may miss other etiologies that explain the clinical manifestations, such as a separate inherited prothrombotic risk factor (e.g., FVL) in a family member with venous thromboembolism. Several families have been reported with aPL combined with a second hematologic defect, like FVL [14], factor XII deficiency [15], and a factor IX inhibitor [16], with additional data supporting a risk synergy between classic inherited thrombophilias and APS.
Multiple families have been described in which two or more members have APS. Most of these family investigations are small, however, with details obtained for the affected family members and relatively limited information available concerning unaffected individuals [17–24]. Families with members who have aPL and some of the less obvious clinical manifestations associated with these autoantibodies, such as thrombocytopenia and/or cardiac valve disease, have been described [14, 25].
Another clinical phenotype approach to search for inheritance patterns is to determine the frequency of elevated aPL levels in family members of patients with APS or autoimmune disease [20, 25–28]. Such studies suggest a familial propensity but underscore difficulties in validation of clinical data and likely underdiagnose APS and underestimate its prevalence both in proband families and in control populations [29]. Because spouses of SLE patients may have abnormal LA tests, such studies also demonstrate that environmental factors may play a role in the development of aPL [30].
Sneddon syndrome is livedo reticularis and cerebrovascular ischemic lesions, frequently in association with aPL [31]. Multiplex families with the syndrome and aPL have been described [32–34], including a large family with several individuals with strokes at an early age [35]. In contrast, in at least one family with familial Sneddon syndrome, the clinically affected individuals did not have aPL, suggesting that Sneddon syndrome, at least in some cases, is a separate clinical entity from APS [36].
Genotype Evaluations
While genetic analyses related to nonfamilial APS are frequent, few exist that evaluate families. Goel and colleagues reported the first large-scale genetic study of multiplex APS families [37]. Their segregation analysis of seven families, with 30 of 101 family members with APS, suggested either a dominant or codominant model for disease inheritance. It failed, however, to find linkage to human leukocyte antigen (HLA) and other candidate genes including β2GPI, antithrombin, factor V, and Fas. A limitation of this study is that the clinical diagnostic criteria for the syndrome were based on a semiquantitative scoring index that differed from the subsequently developed [38] and revised International APS Classification Criteria [39], and likely identified certain individuals as having APS who would not meet current diagnostic criteria.
HLA Associations
A hallmark of autoimmune conditions is the strong association of many of these diseases with genes in the major histocompatibility complex (MHC) region. Many HLA antigens have been associated with aPL, primary APS (PAPS), and APS associated with other autoimmune diseases (Tables 4.2 and 4.3), producing a complicated and confusing dataset for APS. Many of these reports are small case series, occasionally even single multiplex families, and testing for aPL is frequently incompletely documented (e.g., single test performed, positive cutoff values not provided, and positive results not repeated to confirm). Confirmation of meeting classification criteria for APS is also limited as many of the studies were performed before the existence of criteria. Some of these studies also lack, or fail to specify, appropriate ethnic- or gender-matched controls for the populations studied, where gender or ethnicity might account in part for the frequency of associations.
Table 4.2
Positive associations between human antiphospholipid syndrome populations and human leukocyte antigen alleles
Model | Statistically significant findings based on multivariate analysesa |
|---|---|
SLE population studies | aCL: DRB1*0402 (DR4), DR7 SAPS: DR7 Worsened survival: DQw7 |
SS population study | aCL: DR2, DR3 (Note in other settings, DR2 has been negatively associated with aβ2GPI production in APS) |
APS (both primary and secondary) population studies | LA: DQB1*06 (DQ6), DQB1*0301 (DQw7) aCL: DRB1*0402 (DR4), DR7 Anti-β2GPI: DQA1*03, DQB1*0302 (DQ8), DQA1*0401, DQB1*0604/5 (DQ6, in African Americans), DQB1*0604/5/6/7/9-DQA1*0102-DRB1*1302 haplotype APS: DR7, DMA*0102 |
PAPS population studies | PAPS: DR5 (DRB1*1201), DRw53, DQ7 PAPS and aβ2GPI: DQB1*0604/5/6/7/9-DQA1*0102-DRB1*1302 haplotype |
Table 4.3
Human leukocyte antigen associations reported for nonfamilial and familial antiphospholipid syndrome
Author | Ethnicity/location | Patients (n) | Control (n) | Phenotype associations evaluated | Results |
|---|---|---|---|---|---|
Nonfamilial | |||||
Panzer et al. [97] | Austrian | 27 mixed (PAPS, 22; SAPS due to SLE, 4; SLE only, 1) | 637 | aCL, LA, antiplatelet antibodies | Increased frequency of HLA-DQB1*06 with LA after correction for multiplicity testing |
Freitas et al. [98] | Brazilian | 123 mixed (PAPS, 34; SAPS due to SLE, 35; SLE only, 54) | 166 | APS (by criteria) and SLE | PAPS with nonsignificant increased frequency of DR53-associated alleles; association of SAPS with HLA-DRB1*03 was due to the association with SLE and not aCL and suggested that the HLA class II profile of PAPS is different from that of SAPS |
McHugh et al. [99] | British | 46 SLE | 318 | aCL | DR4 in 7/8 aCL-positive SLE patients; associations of NS after multiplicity testing |
Asherson et al. [100] | British | 13 PAPS | 69 | aCL, LA, PAPS | DRw53 positively associated with PAPS and aCL; DR4 positively associated with PAPS; DR3 not present in any patients vs controls; associations NS after multiplicity testing |
Sanchez et al. [101] | British Caucasian | 133 mixed (PAPS:51; SAPS due to SLE:42; SLE only:40) | 109 | any aPL, aCL, aβ2GPI, aPT, LA | The distribution of DMA alleles was significantly different between all APS patients or SAPS patients and controls with the increase in DMA*0102 showing the strongest contribution. The distribution of DMA alleles in PAPS or SLE only patients was not significantly different from that in controls |
Bertolaccini et al. [102] | British Caucasian | 82 mixed (all with aPL and 74 with APS: PAPS:53; SAPS due to SLE:29; SAPS due to other: 2) | 177 | aβ2GPI, LA, aPS/PT | HLA-DQB1*0301/4 associated with aPS/PT with or without aβ2GPI with increased frequency. Uncertain if multiplicity corrections applied |
Caliz et al. [103] | British Caucasian | 83 mixed (PAPS:53; SAPS due to SLE:30) | 177 | aβ2GPI, APS features | DQB1*0604/5/6/7/9-DQA1*0102-DRB1*1302 and DQB1*0303-DQA1*0201-DRB1*0701 haplotypes showed significantly positive correlations with APS that were NS after correction. The association of the former was significant after correction only in PAPS with aβ2GPI. |
Goldstein et al. [104] | Canadian | 107 mixed (PAPS:16; SLE with aPL:19; SLE without aPL:72) | aCL, LA, APS | HLA-DR4 and the linked DR53 were significantly increased in PAPS compared to SLE. In patients with aPL (SLE and PAPS) compared to patients with SLE without aPL, associations were found with HLA-DR53 and to a slightly lesser degree DQ7. The HLA-B8, DR17, DQ2 haplotype closely associated with SLE was significantly decreased in both SLE with aPL and with PAPS. No evidence of association with C4A deficiency alleles. No correction for multiplicity testing appears to have been applied | |
Hartung et al. [105] | Central Europe | 314 SLE (SAPS:17) | – | aCL IgM and IgG, APS | aCL IgM were positively associated with DR4, DR7, DRw53; DR7 association NS after corrections for multiplicity testing; no evidence for significant associations with DQ or C4 alleles or for aCL IgG or APS. |
European | 577 SLE | Unknown | aCL, aβ2GPI, APS features | aCL were positively associated with HLA-DRB1*04, -DRB1*07, -DQA1*0201, -DQA1*0301, -DQB1*0302, -DRB3*0301, -DPB1*1501, and -DPB1*2301. aβ2GPI were positively associated with HLA-DQB1*0302, -DPB1*0301, and -DPB1*1901. HLA-DQA1*0501 and -DRB3*0202 showed a negative association with aCL. Although aCL and aβ2GPI were associated with HLA-DRB1*0402 and -DRB1*0403, only the -DRB1*0402 association was still positive with med-high titer aCL after correction for multiplicity testing. No associations were found with disease manifestations after correction | |
Ioannidis et al. [108] | Greek | 67 mixed (PAPS:37; SAPS:30) | 246 | aβ2GPI | aβ2GPI response was positively associated with HLA-DQA1*03 (in particular *0301) and the HLA-DRB1*1302-DQB1*0604 haplotype, while protection against developing an aβ2GPI response was related to the HLA-DRB1*0101-DQA1*0101 haplotype and the HLA-DRB1*1101 allele. Correction for multiplicity testing doesn’t appear to have been applied |
Savi M et al. [109] | Italian | 109 mixed (PAPS:19; SLE with aCL:36; SLE without aCL:44) | 2 groups: 319 and 633 | aCL positively associated with DR7; DR2 increased among aCL(−);uncertain if multiplicity corrections applied | |
Sebastiani et al. [110] | Italian | 44 SLE | 100 | IgG or IgM aCL | No significant associations with DR alleles found |
Trabace et al. [111] | Italian | 49 women randomly chosen from 120 with RPL (aCL[+]:25; aCL[-]:24) | 100 including 54 men | aCL | HLA-DR7 in aCL(+) vs aCL(−) (NS after adjustment for multiple comparisons) but no differences vs healthy controls |
Hashimoto et al. [112] | Japanese | 145 SLE (29 with aβ2GPI) | 113 | aβ2GPI | The frequency of DRB1*0901 (DR9) was lower in SLE patients than in healthy subjects. SLE patients with aβ2GPI-showed significant positive association with DRB1*0901 compared to those without aβ2GPI; associations NS after correction for multiplicity testing |
Vargas-Alarcon et al. [113] | Mexican | 17 PAPS (all with aCL) | 100 | PAPS | Significant positive association of HLA-DR5 with trend for DR5 subtype of -DRB1*1201. Increased frequency of DR52 also noted (NS). No associations seen with DQ7, DR53, DR7 |
Granados et al. [114] | Mexican | 80 SLE | 378 first-degree relatives; 50 married couples without SLE | aCL, APS | After multiplicity correction, positive association with aCL and APS noted with DR7 |
Camps et al. [115] | Southern Spain | 19 PAPS | 261 | aCL and LA, APS features | DQ7 and DRw53 associated with PAPS after multivariate analysis. DR4 numerically positively associated with high-titer IgG aCL, whereas DR7 was associated with low- or medium-titer IgG aCL. No HLA associations with LA. DQ1 significantly positively associated with migraine; DQ6 with RPL; DR1 with HTN and epilepsy (multiplicity corrections do not appear to have been applied to APS feature analyses) |
Wilson et al. [116] | US: AA | 44 SLE | 38 | aCL | A statistically significant association with IgG aCL and C4-null allele was seen. No correction for multiplicity testing was performed |
Arnett et al. [117] | US | 20 with LA (PAPS, 8; SLE, 9; SS, 2; SSc, 1) | 139 | LA | HLA-DQw7 (DQB1*0301) linked to HLA-DR4 or DR5 significantly associated after correction in all LA(+) patients and SLE LA(+) patients; among the HLA-DQB1 *0301 (DQw7)-negative patients, all possessed HLA-DQw8 (DQB1 * 0302) and/or HLA-DQw6 (DQB1 *0602 or DQB1 *0603) alleles |
Asherson et al. [118] | US | 65 SS (5 males) | 150 white women | aCL (including IgA) | Significant positive association after correction between aCL and HLA-DR2/DR3—these were also associated with anti-Ro/SSA. No increased occurrence of haplotype DR2 or DR3 was noted for the SS patients vs controls, suggesting that gene interaction between DR2 and DR3 may play a part in the production of aPL in SS patients. Of the 13 subjects with aCL, 11 had IgA, 1 IgM, and 3 with IgG aCL. None had high titer IgM or IgG aCL. No subject had LA |
Arnett et al. [119] | US: 3 ethnic groups (white, AA Mexican-American) | 262 mixed (PAPS, 48; SAPS due to SLE, 70; SLE only, 126; other SAPS, 4; other CTD, 14) | 393 | aβ2GPI | After correction for multiplicity, in whole patient group: DR2, DRB1*1501 and/or 1503 (DR2), and DQB1*0602 (DQ6) negatively associated with aβ2GPI; DQA1*03, DQB1*0302, and DQA1*0401 positively associated with aΒ2GPI. In assessing aβ2GPI vs controls: DQA1*0101 (DR1) negatively associated and DQB1*03 and DQB1*0302 (DQ8-linked to DR4) positively associated with aβ2GPI. In AA patients, DQB1*0604 or 0605 positively associated and DQB1*0602 negatively associated with aβ2GPI. In Mexican Americans, DQA1*03 positively associated with aβ2GPI |
Galeazzi et al. [120] | Unknown | 42 SLE | 107 | aCL | DPB1*0301 and/or DPB1*1401 after correction were statistically significantly increased in aCL-positive but also Sm/RNP-positive patients compared with 107 healthy controls; this was also statistically significant in aCL-positive versus aCL-negative patients, but not maintained after correction |
Gulko et al. [121] | US, mixed ethnicities and races | 139 SLE | None | aCL, APS features, survival | HLA-DQw7 and thromboembolic events statistically significantly and independently in multivariate analyses adversely affected survival. No HLA association of DR or DQ alleles with aCL found |
Familial | |||||
Bhattacharya et al. [122] | British | 8 members of a single family, 2 with APS, one with lupus nephritis | – | LA | The proband and father had APS; a paternal aunt had lupus nephritis. Five members displayed LA; none had aCL except the proband. Six members shared the same haplotype, A30, B13, DRB1–07 (DR7), and DQB1–02 (DQ2): four of these had LA. |
Dagenais et al. [123] | Canadian, English | 14 members of a single family, some with AID and/or aPL; proband had APS | – | aCL, LA | The proband and 4 other family members shared the HLA-B60, DR4 haplotype; of these the proband had APS and 3 others were aCL positive without symptoms. No one without the haplotype had aCL |
Hudson et al. [124] | Canadian, Native American | 8 members of a single family, 4 with aCL | – | aCL | 3 of 4 with aCL had DRB1*14, but 1 nonaffected family member did also |
Bridey et al. [125] | French | 13 members of a single family, 2 including proband with PAPS | – | aCL, LA, aβ2GPI | Haplotype A11B51 C4A3BQ0 DR4 DRw53 DRB1*0402 was shared by proband and affected brother and seven other family members. Proband and brother were documented to have LA and aCL; proband also had aβ2GPI. Two additional asymptomatic members with the haplotype had aβ2GPI. Three of the 4 asymptomatic family members with borderline LA results shared the haplotype |
Lousa et al. [126] | Spanish Caucasian | 19 members of a single family, 13 HLA-typed; 2 with PAPS including proband who also had Sneddon syndrome, 1 with livedo reticularis; 10 had LA and/or IgM aCL | – | aCL, LA | The proband and father (as well as 1 asymptomatic nephew negative for both aCL and LA) had an HLA-A30-B13-Bw6 haplotype. In addition, an HLA-Bw6-DQ1 association was present in all the typed members of this kindred |
May et al. [127] | US | 8 members of a single family, 3 affected (mother and identical twins) with SAPS due to SLE | – | aCL, LA, APS features | aCL and LA present in all affected subjects with haplotype of DR4, DRw53, and DQw7, but haplotypes also present in a nonaffected brother and nonaffected sister; no evidence of C4A or C4B deficiency |
Goel et al. [37] | US | 101 members, 30 affected from 7 families | – | APS | No HLA associations found |
Wilson et al. [128] | US Caucasians | 38 members, 19 with AID (including PAPS but not SLE) or autoantibodies from three families | 33 | aCL | In ten members with aCL, all had 1 or 2 DQB1 risk alleles, and 9 of 10 had 1 or 2 C4 deficiency alleles |
Non-HLA Associations
β2-glycoprotein I is a phospholipid-binding protein that has been identified as a major antigen in patients with APS. The protein sequence for this plasma protein was published in 1984 [40], and the complementary DNA sequence in 1991 [41, 42]. Several polymorphisms have been identified in the protein [43], including three in the phospholipid-binding fifth domain (Val/Leu247, Cys/Gly306, and Trp/Ser316). Studies have investigated the relationship between polymorphisms in β2GPI and anti-β2-glycoprotein-I (aβ2GPI) in various ethnic groups. Gushiken and colleagues found no association between aPL (LA or aβ2GPI) and the Cys/Gly306 and Trp/Ser316 polymorphisms which disrupt the ability of the protein to bind to anionic phospholipids [44], in patients with SLE and/or APS [45]. Similarly, Camilleri and colleagues found no relationship between the Trp/Ser316 polymorphism and aPL in patients with thrombosis [46]. Palomo and colleagues did find a significant relationship between this polymorphism and venous and arterial thromboses, but not with aPL, in Chilean patients [47]. Similar findings were reported by Pardos-Gea et al.; i.e., polymorphisms in Trp/Ser316, by means of statistically significant associations for an increased S allele and T/S genotype in Spanish Caucasian patients, might play a role in the pathogenic development of PAPS, but not via increased production of aβ2GPI or other aPL [48].
The Val/Leu247 polymorphism has been more extensively studied. This variant causes a conformational change in β2GPI [49] hypothesized to result in the exposure of cryptic epitopes [50], theoretically providing a likely target for autoantibodies. Initial studies identified a relationship between this polymorphism and aβ2GPI in patients with APS, although one study identified this association in Caucasian patients with primary but not secondary APS [51], and a second study identified the association in Asian patients with APS but not Caucasian or African American patients [52]. To evaluate not only potential differences related to ethnicity or the presence of primary versus secondary APS, a recent meta-analysis by Chamorro et al. of eight previous studies, comprising 488 patients meeting classification criteria for APS and 923 controls from eight countries, evaluated the association of APS, PAPS, aβ2GPI, and/or thrombosis with the Val/Leu247 polymorphism [53], determining that patients with APS had a significantly higher prevalence of the Val/Val genotype when compared with controls, with an enhanced association in those with aβ2GPI. No associations were found for the Val/Val phenotype and thrombosis, ethnicity (Caucasians with APS), or PAPS, but these analyses were limited by the small number of studies reporting these data. A separate meta-analysis evaluating the Val/Leu247 polymorphism included 10 studies with a total of 1507 patients and 1450 controls [54]. A limitation of this meta-analysis was that it did not confirm that patients met classification criteria for APS as done in the Chamorro meta-analysis. Only six studies were common to both meta-analyses. Despite the differences, the latter study confirmed the findings of the former except the latter also demonstrated an association of the polymorphism with thrombosis.
Several other genes have been studied in focused attempts to identify relationships with aPL and APS (Table 4.4). Fredi and colleagues studied 169 Italian patients with PAPS for polymorphisms in non-HLA genes that are associated with increased susceptibility for lupus, including IRF5, STAT4, and BLK [63] and found a strong genetic association with PAPS for STAT4 and BLK and a weak association for IRF5. In a similar study, 48 single nucleotide polymorphisms (SNP) from 40 candidate loci for lupus were typed in a cohort of 208 Italian and Hungarian SLE patients and 152 controls [65]. No associations between secondary APS and IRF5, STAT4, or BLK were found; weak associations that did not persist after multivariate analysis were found with NCF2 and TYRP1. In contrast, in separate studies, a STAT4 polymorphism was associated with primary as well as secondary APS [58], and a different STAT4 polymorphism was associated with aPL and ischemic cerebrovascular events in Swedish patients with lupus [59]. These conflicting data suggest that while genetic variants that increase the risk for the development of PAPS may overlap with those that increase the risk for the development of SLE, larger studies are needed to determine both those genes which truly contribute to heritable risk and why different disease phenotypes manifest.
Table 4.4




Non-HLA genetic associations for human antiphospholipid syndrome populations from non-genome-wide studies
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree