Fig. 1
Flowchart of processes in the acquisition of specimens prior to sample analysis. This study focuses on sampling of tissue specimens fresh-frozen with entry at pathology
Biorepositories, tissue, and sample collections of archived patient materials that allow to initiate and to evaluate retrospective and prospective clinical studies, to study biomarkers or to address pharmacokinetic questions [12], heavily rely on sample material of high quality. Apart from preparation and storage, sample quality is primarily ensured by processing of tissues of best possible quality [4]. Mainly used for expression studies, RNA is distinguished from other cellular components by its low stability. For scientific purposes, the preservation of native RNA in surgical specimens is of utmost importance and various approaches have been taken to minimize RNA degradation processes [10].
The introduction of RNA integrity numbers (RIN) greatly facilitated the discrimination of native versus degraded RNA [11]. Based on integrity measurement of S18 and S28 ribosomal RNAs, the determination of RIN values can be utilized as a quality standard for fresh-frozen tissue at the molecular level [3].
One approach to preserve RNA integrity is fresh-freezing of tissue specimens prior to preparation. Vice versa, assessment of RNA integrity can be utilized as a quality indicator for sample preservation [13] to answer the questions: how does tissue type influence RNA quality in fresh-frozen samples and does processing time in a pathological laboratory affect RNA quality? A recent approach to ensure tissue preservation is vacuum-sealing of fresh tissues [2], here compared with fresh-frozen tissues of specimens of the gastrointestinal (GI) tract in particular with RNA integrity as the quality indicator.
The aim of this study was quality assessment of various pre-analytical steps with special focus on sample preservation of GI specimens. RNA integrity expressed as RIN values served as the quality indicator to evaluate the impact of organ sites and processing times in the pathology laboratory on sample quality.
2 Methods
Fresh-frozen tumor tissue samples, collected between 2009 and 2012 at the Institute of Pathology at the University Hospital Carl Gustav Carus, Technische Universität Dresden, and stored at −80 °C and or in the gaseous phase of liquid nitrogen, were randomly selected (n = 171) to generate a representative sample cohort.
RNA extraction was performed using the RNeasy microAmpKit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. RIN (RNA integrity number) values were determined on the RNA-6000-Nano-LabChip-Kit with the Agilent-2100-Bioanalyzer (Agilent Technologies, Böblingen, Germany). Extracted RNA was used at a concentration of 25 ng/μl.
Statistical analysis was performed using the SPSS software (IBM, Ehningen, Germany). Sample size was estimated with nQuery Advisor® 7.0 (Statistical Solutions Ltd., Cork, Ireland).
3 Results
3.1 Integrity of RNA Derived from Fresh-Frozen Tissues of Different Organs Varies Substantially
Overall, 171 fresh-frozen tissue specimens stored at −80 °C or at −196 °C in liquid nitrogen were randomly collected to create a representative sample for quality assessment by RNA extraction and evaluation of integrity with the RIN algorithm. The collection is composed of specimens of lung, breast, stomach, liver, pancreas, kidney, small intestine, colon, and rectum as listed in Table 1. RIN values were classified as of sufficient quality (RIN ≥ 5) or as inadequate for further analyses (RIN < 5) according to the literature [11]. Primary analysis indicated that RNA integrity from tissues derived from the gastrointestinal tract was on average of significantly lower quality compared to other organs (Fig. 2, ***, p < 10−3) and a smaller fraction of fresh-frozen tissue samples derived from GI specimens was suitable for subsequent studies compared to other organs. Here, about 70 % of samples derived from breast and kidney specimens were suited for further analysis (RIN ≥ 5), whereas lung specimens rarely produced samples of sufficient RNA quality (1/6 specimens). A further grading of suitability for sample derivatization is observed in the group of GI organs (highlighted in green in Table 1). In the majority of GI organs, about half of specimens appear suitable for derivatization, apart from stomach specimens with only 26 % (5/19) of samples with sufficient RNA quality.
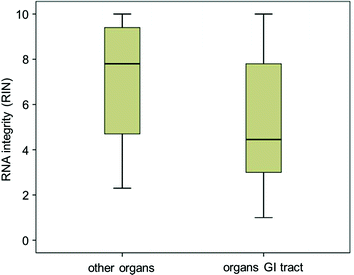
Table 1
Tissue origin affects RNA integrity of fresh-frozen tissue samples
Tissue origin | Specimens (n) | RIN ≥ 5 (%) | RIN < 5 (%) | ||
|---|---|---|---|---|---|
liver (Li) | 32 | 17 (53) | 15 (47) | ||
Colon (C) | 27 | 13 (48) | 14 (52) | ||
Rectum (R) | 25 | 10.5 (42) | 14.5 (58) | ||
Breast (B) | 22 | 16 (73) | 6 (27) | ||
Stomach (St) | 19 | 5 (26) | 14 (74) | ||
Kidney (K) | 19 | 13 (68) | 6 (32) | ||
Pancreas (Pa) | 13 | 5.5 (42) | 7.5 (58) | ||
Lung (Lu) | 6 | 1 (17) | 5 (83) | ||
Small intestine (SI) | 4 | 3 (75) | 1 (25) | ||
Prostate (Pr) | 4 | 3 (75) | 1 (25) | ||
Total | 171 | 87 (51) | 84 (49) | ||

Fig. 2
Diminished RNA preservation in samples derived from organs of the gastrointestinal (GI) tract compared to other organs (p < 10−3). Box plot analysis of RIN values of GI versus other tissues. Results of organs of the GI tract were grouped and compared to RIN values of other organs listed in Table 1
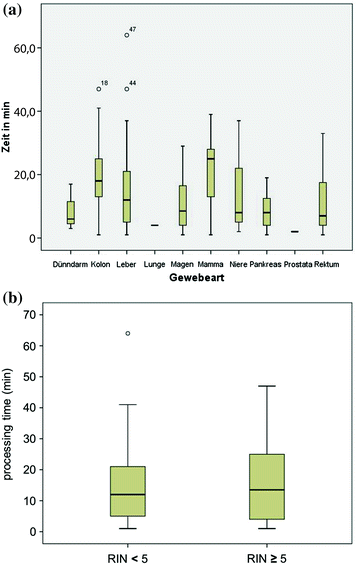
3.2 Processing of Tissue Specimens Does Not Contribute to Tissue-Specificity of Sample Quality
Ignoring intrinsic factors that affect RNA integrity like pancreatic enzymes, gastric acids, or bacterial colonization of colon and rectum, our work focused on processes suitable for optimization of tissue specimens in the molecular pathological laboratory. Sample processing times are closely linked to RNA integrity, and analysis of sample processing times after entry at pathology reveal tissue-specific differences in times to freezing of specimens (Fig. 3a). However, processing times did not differ (p = 0.502) between samples of sufficient RNA quality (RIN ≥ 5) compared to samples with lower RIN values (<5). Thus RNA integrity as indicator of sample quality is not affected by processing times (Fig. 3b) of tissue specimens at pathology. Taken together, these results show that even though organ-specific variations of processing times in pathology do occur, these variations do not contribute to sample quality as evaluated by RNA integrity as a quality indicator.