Fig. 10.1
Oncogenic activation of mTOR may be related to multiple oncogenic activations, mutations of major anti-oncogenes as well as microenvironment-dependent parameters such as hypoxia and nutrient depletion
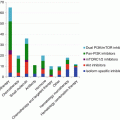
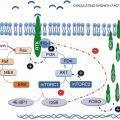
Knowing how the mTOR pathway is activated and negatively regulated is crucial to understand which biological settings might represent potential candidates for treatment with mTOR inhibitors or conversely which tumor types may be primary or secondary resistant to rapalogues. Moreover, since the PI3K-AKT-mTOR pathway is activated both in endothelial and tumor cells, the overall effects of rapamycin derivatives may vary according to solid tumor addiction upon cell survival and/or angiogenesis [8, 9]. Whereas certain biological parameters, such as S6K1 activity, can reflect the exposure to rapamycin derivatives [10], we will restrict this article to the question of predictive biomarkers, determined at the baseline prior to the initiation of rapamycin-based therapy. High-throughput screening for oncogenic events and epigenetic changes occurring in tumors from individual patients are likely to become essential tools to optimize the use of drugs inhibiting mTOR functions.
10.4 Sensitivity to mTOR Inhibitors and Activation of the PI3K-AKT Pathway
The identification of tumor types that may respond to mTOR inhibitors remains a major issue. Since mTOR is ubiquitously expressed in tumor tissues and healthy organs, the sensitivity or resistance to mTOR inhibitors cannot be predicted upon the presence or absence of the target. For this reason, the overall activation of the PI3K/AKT/mTOR pathway has been proposed to identify tumor types that could be sensitive to rapalogues. However, thus far, parameters reflecting activation of the PI3K/AKT/mTOR pathway have failed to predict sensitivity to rapalogues in most tumor types. Main intrinsic parameters of this pathway that have been assessed in tumor models as biomarkers of sensitivity, alone or in combination, have been the loss of PTEN function, AKT phosphorylation, and PI3K mutations.
Neshat et al. [11] first reported the enhanced sensitivity of PTEN-deficient tumors to the inhibition of mTOR. Using several cell lines of glioblastoma and prostate and breast cancers, the authors showed that PTEN-null cells were more sensitive to the rapamycin derivative temsirolimus than PTEN wild-type cancer cells. This was confirmed in vivo by using human prostate xenografts, against which temsirolimus displayed limited activity when PTEN was functional, requiring high doses to achieve antitumor effect. In contrast, temsirolimus showed significant growth inhibition in PTEN-null xenografts, even when using relatively low concentrations [6]. Since PTEN inactivation is often associated with poor outcome, PTEN inactivation corresponding to mutation and loss of protein expression has also been described in sporadic tumors such as glioblastoma and endometrial, prostate, and breast cancers, as well as melanoma, making those tumors theoretically candidates for treatment with mTOR inhibitors [12–14]. In addition, mTOR inhibition was shown to reverse doxorubicin resistance conferred by PTEN status in prostate cancer cells [15]. However, thus far, no correlation between PTEN expression and clinical activity was demonstrated in clinical trials, and therefore lack of PTEN expression in cancer cells from patient biopsies cannot be reliably used to select patients candidate to mTOR and/or PI3K inhibitor treatments.
Another biomarker of responsiveness to mTOR inhibitors suggested by other authors was the level of phosphorylated AKT (p-AKT) in cancer cells. As an example, high p-AKT level and reduced PTEN expression rendered renal carcinoma cell lines potentially sensitive to mTOR inhibition [16]. However, several recent papers have further argued against the significance of AKT activation. For example, a high level of p-AKT might not only reflect the activation status of the pathway induced by upstream signals but may also result from the feedback loop induced by mTORC2 (mTOR-RICTOR), characterized by the rapamycin-insensitive mTOR activity [17]. As it stands, it would be therefore interesting to explore whether the number of RICTOR copies could predict resistance to rapamycin derivatives. To our knowledge, this parameter has not yet been investigated in clinical situations. Furthermore, AKT activation has been reported to be associated with development of cell resistance to rapalogues [18], to conventional cytotoxics [19], and to EGFR inhibitors in tumors cells displaying a mesenchymal phenotype [20]. For these reasons, high levels of phosphorylated AKT in tumor cells might not be used as a predictor of response to mTORC1 inhibitors but rather be regarded as a determinant of resistance to a broad variety of anticancer agents, including rapamycin derivatives. Although preclinical data suggested that AKT activation was associated with sensitivity and/or resistance to PI3K and mTOR inhibitors, expression of AKT cannot be recommended outside clinical trials to select patients for therapeutic interventions.
More recently, activating mutations affecting the catalytic subunit of PI3K (p110α encoded by PIK3CA gene) were reported [21]. The occurrence of such mutations may reach 25–30 % of sporadic epithelial tumors, including breast, colon, prostate, and endometrial carcinomas. The potential correlation between PI3K status and sensitivity to mTOR inhibitors has been less extensively described. In a recent publication, Di Nicolantonio et al. [22] showed that two different cell lines of breast cancer harboring p110α-activating mutation (PIK3CA mutation) had increased sensitivity to everolimus as compared to their wild-type PI3K counterparts. However, those data are preliminary, and the PIK3CA-driving mutation hypothesis shall now be further evaluated in larger clinical trials.
In addition to the abovementioned biological parameters, malignancies such as mantle cell lymphoma are potential candidates for treatment with rapalogues because cyclin D1 mRNA overexpression primarily drives these tumors. The translational regulation of cyclin D1 is under direct dependency of PI3K/AKT/mTOR pathway. Given the role of cyclin D1 in mantle cell lymphoma, several trials in relapse setting have demonstrated the effects of temsirolimus, further confirming that cyclin D1 overexpression appeared to be predictive of sensitivity to mTOR inhibitors in this disease [23, 24]. In renal cell carcinoma, the investigators involved in the global phase III study comparing temsirolimus to interferon alpha have conducted exploratory analyses to determine if the molecular markers PTEN and hypoxia-inducing factor (HIF)-1α were correlated with efficacy. Figlin et al. [25] reported that the baseline status of PTEN and HIF-1α did not correlate with efficacy in renal cell carcinoma patients treated with temsirolimus versus IFN. In this study, patients demonstrated overall survival and progression-free survival benefit when treated with temsirolimus regardless of PTEN and HIF-1α status. The authors concluded that baseline PTEN and HIF-1α levels might not be used to predict response to temsirolimus in patients with advanced renal cell carcinoma.
A number of other potential biomarkers were explored in endometrial tumor samples, including phosphorylated S6, phosphorylated 4E-BP1, hTERT, and telomere length, but none were found to be effective in discriminating which tumors would best respond to the antiproliferative effects of rapamycin treatment [26].
In summary, preclinical studies mainly based upon in vitro cultured tumor cell lines suggest that the effects of mTOR inhibitors may be more pronounced in cancers displaying loss of PTEN function or PIK3CA mutations. However, this statement does not readily translate in clinical settings [27]. This is well illustrated by a recent paper searching for biomarkers of sensitivity, using freshly expanded endometrial cancers from surgical specimens to evaluate the effect of rapamycin [26]. The authors characterized the explants regarding wild-type PTEN and p-AKT status by using western blotting. Among 13 cases, 7 cases displayed expression for wild-type PTEN and 12 other samples showed p-AKT. Using a short-term culture assay, 9/13 specimens responded to rapamycin, with a median IC50 of 11.4 nM (range 0.01–50 nM). Rapamycin inhibited cell growth both in PTEN-positive (5/7) and in PTEN-negative (4/6) surgical specimens of endometrial cancers. Although limited to small numbers, this study suggests that sensitivity to rapamycin is neither exclusively dependent on the functionality of PTEN nor of the AKT phosphorylation status. Other works using breast cancer and glioblastoma cell lines have found that loss of PTEN function is insufficient as a single parameter to predict response to mTOR inhibitors both in vitro and in vivo [28, 29].
10.5 Molecular Biomarkers of Resistance to mTOR Inhibitors
10.5.1 Bcl-2 Overexpression
In cancer and endothelial cells addicted to the PI3K/AKT/mTOR pathway and with functional apoptosis, relatively low doses of rapamycin derivatives might be sufficient to induce cell death. This may explain why antitumor activity in the clinic was not fully dose dependent and why objective responses were observed sporadically with rapamycin derivatives in several malignancies such as renal cell carcinoma [30, 31], gastrointestinal neuroendocrine tumors [32, 33], and mantle cell lymphoma [23, 24].
Conversely, in tumors that are marginally sensitive or resistant to mTOR inhibitors, higher doses of rapamycin derivatives may be necessary to induce cell death. Another limiting key factor of the resistance to rapamycin is that tumor cells may have nonfunctional apoptotic pathway, especially when expressing Bcl-2, which remains a major protein involved in resistance to apoptosis. Illustrating this paradigm, our team has shown that rapamycin-resistant SKOV3 ovarian cancer cells (harboring a functional PI3K/AKT pathway) overexpressed the apoptosis-inhibitory protein Bcl-2 as compared to IGROV1-sensitive cells. To determine the specific role of Bcl-2, we used BCL2 antisense oligonucleotides designed to interact with BCL2 mRNA. This strategy was able to restore the apoptotic response to everolimus in SKOV3 tumor cells. This study demonstrated that Bcl-2 had a critical role in preventing apoptosis induced by rapamycin derivatives [34]. Other data are consistent with our findings in mice bearing transgenes encoding both AKT and Bcl-2, in which prostate intraepithelial neoplastic cells remained sensitive to RAD001-induced inhibition of proliferation but were resistant to apoptosis [35].
Taken together, the above results suggest that overexpression of antiapoptotic proteins such as Bcl-2 might serve as a surrogate marker for resistance to rapalogues. However, it remains to be shown whether expression of Bcl-2 and its homologues (such as BCL-XL, BCL-w) could predict resistance to mTOR inhibitors in the clinical setting. These findings motivate the development of synergistic combinations between rapalogues and classical cytotoxics, with the aim to restore apoptosis in tumor cells. Our team, along with others, has investigated such combinations in preclinical models [36, 37]. By using three different head and neck cell lines, we have shown synergistic effects when rapamycin was combined with carboplatin or paclitaxel, the most active sequence being chemotherapy followed by rapamycin. Looking at cell cycle effects, we found that the choice of the sequence might be important to optimize efficacy, the induction of apoptosis being far more pronounced with chemotherapy followed by rapamycin, in comparison to the opposite sequence [36]. The poor results observed with rapamycin followed by chemotherapy may be explained by rapamycin-induced G1 arrest that may not allow chemotherapy agent to exert its optimal antitumor effect, especially if the agent is active in S or M phase of the cell cycle. Our team recently completed a prospective phase I–II trial in patients with advanced head and neck carcinoma, investigating the tolerance and efficacy of rapamycin combined with carboplatin and paclitaxel, given on a weekly schedule as induction chemotherapy prior to radiation therapy [38].
10.5.2 KRAS Mutation May Drive Resistance to mTOR Inhibitors
Another hypothesis yielding to cell survival despite inhibiting mTOR is the presence of alternative survival pathways. To maintain survival and proliferation, tumor cells might be using redundant transduction pathways, involving particularly the MAPK signaling. Di Nicolantonio et al. [22] previously underlined the strong impact of KRAS mutations that was shown capable of overcoming the inhibiting effects of everolimus on the PI3K/AKT/mTOR pathway. In the first part of their work, the authors demonstrated that introducing PIK3CA mutation sensitized breast cancer cells to the effects of everolimus in comparison to parental cells (cf. supra). In the second part of this publication, by treating a panel of cell lines derived from glioblastoma and breast, ovarian, prostate, endometrial, and colorectal carcinomas, they reported that everolimus-resistant cells (such as HT-29, HCT116, and DLD-1) carried mutations in both PIK3CA and KRAS/BRAF. Furthermore, they elegantly demonstrated that genetic ablation of the KRAS D13 mutation restored the antiproliferative response of cancer cells to everolimus, both in vitro and in vivo. While investigating more in details of the mechanisms of resistance, the authors found that KRAS could activate translation through an mTORC1-independent pathway and therefore could bypass everolimus-mediated mTOR inhibition, possibly through the activation of p90 ribosomal S6 kinase (p90RSK). In this situation, activation of the RAS-ERK1/2 cascade and of RSK1 may provide an alternative route to translational control. Importantly, the authors also investigated their findings in clinical situations by assessing the mutational status of PIK3CA, KRAS, and BRAF in a cohort of cancer patients treated with single-agent everolimus as part of phase I-II studies. They showed that patients whose tumors harbored PIK3CA mutations or PTEN loss of function had increased clinical benefits from everolimus treatment, except when KRAS mutations were present, the latter situation being associated with lack of response by univariate analysis. As such, this study illustrates the importance of KRAS mutations in preclinical models and clinical setting, yielding to circumvent the effects of mTORC1 inhibition by everolimus, through activation of alternative RAS-dependent survival pathways, including MAPK. As it stands, another approach to circumvent mechanisms of resistance to mTOR inhibitors could be to combine such inhibitors to other targeted agents, for example, with MEK inhibitors that have been shown to be active in the case of KRAS mutation [39].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree