Fig. 1
Phylogenetic tree of the HPV16 variants identified by Yamada et al. (1995) in the United States within a sample of 30 HPV16 isolates based on parsimony analysis. Combined sequences from E6, L2, L1, and the LCR for 30 isolates (including the HPV16 reference [REF.] genome) yielded in their analyses an alignment of 129 variable positions. The single most parsimonious tree (157 steps) is shown. Small numbers above branches indicate numbers of steps (reconstructed point mutations) along the corresponding branch; the horizontal length of each branch is proportional to the number of steps, while vertical branch length is for layout only. Large numbers below branches indicate bootstrap values ≥90 %. On the right site, the HPV16 classes are indicated
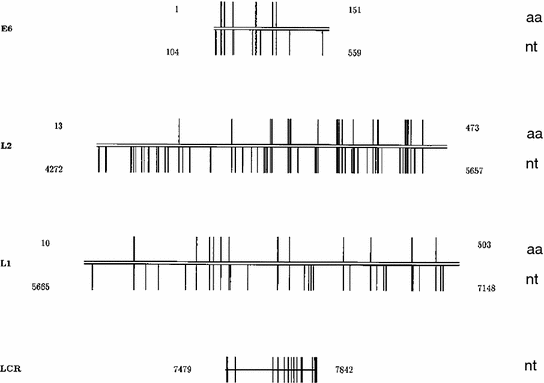
Fig. 2
Distribution of nucleotide (nt) and amino acid (aa) changes among HPV16 within E6, L2, and L1 coding sequences and a segment of the LCR. For the coding regions, the beginning and ending nucleotide positions are indicated below the lines to the left and right, respectively; the beginning and ending residues of the predicted amino acid sequences are indicated above the lines. Vertical bars below the lines represent the positions of nucleotide substitutions, while vertical bars above the lines represent predicted amino acid changes. For the LCR, the beginning and ending nucleotide positions are indicated to the left and the right of the line, while bars through the line represent the positions of nucleotide substitutions (from Yamada et al. 1995, modified)
5 HPV16 Variants Have Variable Carcinogenic Potential
Most findings about variable carcinogenic potential of HPV16 variants come from research on cervix carcinoma. Women with non-E (including Af2 but predominantly AA) HPV16 variants have a 4.5 (95 % CI 1.2–16.8) times higher risk to develop CIN 2/3 lesions than women infected with HPV16 E variants (Xi et al. 1997). In a case-control study, the HPV16 AA variant has also been associated with a higher risk of cervical cancer than E variants; the risk of cervical cancer was eight times higher for patients with AA compared to E variants, when compared to a non-cancer control group (Berumen et al. 2001). Moreover, cancer patients with HPV16 AA were 7.7 years (P = 0.004) younger than patients with the E variant (Berumen et al. 2001). The AA variant was found being the most prevalent HPV16 variant (81.8 %) in Taiwan, and despite the probably expected co-evolution that might have led to adaptation processes, HPV16 AA was also associated with increased prevalence of histologically confirmed CIN grade 3 or worse in women from Taiwan (Chang et al. 2013). Compared to detection of HPV16 E the detection of HPV16 AA was accompanied by an age-adjusted increased odds ratio of 10.7 (1.62–451.05, P = 0.0049; Chang et al. 2013). The same increased risk associated with the AA HPV16 variant was detected in other investigations including prospective trials (Schiffman et al. 2010). However, different oncogenic potential of intra-subtype variants is not limited to HPV16 variants and significant also for other HR-HPV subtypes, e.g. HPV31, HPV35 and HPV51 (Schiffman et al. 2010).
Non-prototype HPV16 variants are associated with higher incidence of cervical neoplasia (Xi et al. 1997). Experimental findings revealed that HPV16 E6 amino acid 83 variants enhance E6-mediated MAPK and differentially regulate tumorigenesis by Notch signaling and oncogenic Ras (Chakrabarti et al. 2004). Related to extensive mutations in the E2 gene the copy number of AA variants per cell is higher than that of E variants (Casas et al. 1999), suggesting that AA variants replicate better than E variants. Since E2 is known to be a regulator of E6 and E7 transcription, this higher mutational level can also contribute to the more aggressive phenotype of AA variants. The European variant T350G, resulting in an amino acid change from leucine to valine at position 83 (L83V) in the E6 protein, is frequently found in cervical intraepithelial neoplasias and cancers and has been associated with progression to cervical cancer particularly in North European women (Zehbe et al. 1998). A recent study from Argentina about HPV16 variants in cervix carcinoma highlighted the most common mutation in the E6 sequences was T350G (L83V), detected in 67 % of the samples, to be associated with increased risk of persistent HPV16 infection (Mosmann et al. 2015). However, the strongest impact on oncogenic activity is caused of the HPV16 AA variant and triggered by their E6 that has some amino acid exchanges promoting cellular immortalization, undergoing transformation to resilient phenotypes, and promoting migration and invasiveness (Niccoli et al. 2012). The E6 protein of HPV16 AA alone is, as shown by in-vitro experiments utilizing keratinocytes solely transfected with the E6 protein of HPV16 AA (Jackson et al. 2014), able to deplete p53 and trigger p16INK4A expression, which is in sharp contrast to E6 from HPV16 E variant.
6 HPV16 Variants in HNSCC
There are only a few reports about HPV16 variants in HNSCC. In a study analyzing 21 HPV16 positive HNSCC from German patients (Hoffmann et al. 2004), only 6 of 21 (29 %) contained the HPV16 prototype sequence, while 8 of 21 (38 %) patients carried a T to G transversion at position 350 (T350G) in the E6 gene, and another 7 of 21 patients (33 %) carried the A131G (R10G in E6) variant together with the C712A mutation (H51N in E7). Since both E6 variants, R10G and L83V, have higher oncogenic potential than the prototype and were found enriched in this small cohort of HPV16-DNA positive HNSCC patients, HPV16 variants might also play an important role in head and neck carcinogenesis. Unfortunately, investigations on HPV16 variants in HNSCC, with the exception of the cited study from Markus Hoffmann and colleagues (Hoffmann et al. 2004), are missing. Nothing is known about their impact on development of HNSCC, treatment response and impact on outcome after treatment. Future investigations on HPV in HNSCC should no longer ignore the potentially very different oncogenic behavior of HPV16 variants that could be one cause for heterogeneity of HPV16 positive HNSCC in their clinical course. Due to geographically distinct patterns of HPV16 variants in cervical lesions (Yamada et al. 1997) and their well-known varying strength in biological effects they are causing, at least some of the differences observed also in the epidemiology of HPV16-driven HNSCC in various regions of the world could be explained. HPV16 positive HNSCC in Asia, Africa, South America and Europe might not be exactly the same as HPV16 positive HNSCC in the United States; they might not be caused by the same HPV16 variants. In the context of the different genetic background of the affected populations also the varying HPV16 variant patterns are expected to be strong modifiers of the respective immune responses to HPV16 infection. Vulnerability for infection with particular HPV16 variants depends on the genetic environment, and persistence of HPV-infection can increase the oncogenic effect of HPV16 and strongly influence the natural course of HPV-driven disease. This might at least partly explain some outcome differences observed in HPV-driven HNSCC. Therefore, investigations on the relevance of HPV16 variants in HNSCC are encouraged.
Acknowledgments
Thanks to all my colleagues and collaborators in the Universities of Leipzig, Gießen, Poznan, Milano, and Heidelberg, the funders of our research, and especially all participating patients and their families.
References
Bernard HU, Chan SY, Manos MM, Ong CK, Villa LL, Delius H, Peyton CL, Bauer HM, Wheeler CM (1994) Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis 170(5):1077–1085CrossRefPubMed
Berumen J, Ordoñez RM, Lazcano E, Salmeron J, Galvan SC, Estrada RA, Yunes E, Garcia-Carranca A, Gonzalez-Lira G (2001) Madrigal-de la Campa A. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J Natl Cancer Inst 93(17):1325–1330CrossRefPubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree