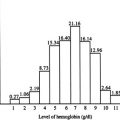
Fetal DNA is present in the plasma of pregnant women and can be used for noninvasive prenatal diagnosis. Early work had focused on the detection of paternally inherited fetal mutations in maternal plasma. Recent advances in single-molecule counting approaches have allowed the mutation dosage of the fetus to be analyzed in maternal plasma. These developments have been demonstrated as feasible for noninvasive prenatal diagnosis of several hemoglobinopathies, including β-thalassemia and hemoglobin E disease.
Prenatal diagnosis is an established part of modern obstetrics practice. Conventional methods for obtaining fetal genetic materials for analysis, however, such as amniocentesis and chorionic villus sampling, are invasive and constitute a risk to the fetus. Over the past few decades, there has been much effort in developing noninvasive methods for prenatal diagnosis that do not have such a risk. Early approaches focused on the isolation of fetal nucleated cells that had entered into the maternal circulation. However, because of the rarity of such cells in the maternal circulation, typically of the order of several cells per milliliter of maternal blood, it has been a challenge to obtain a robust detection of these cells. For example, in a large study designed to investigate the use of these cells for the noninvasive prenatal detection of fetal chromosomal aneuploidies, the detection of circulating male fetal nucleated cells could only be achieved with a sensitivity of 41.4%, with a false-positive rate of 11.1%. As a result of such difficulties, many investigators in the field have moved away from fetal cell isolation to other approaches.
In 1997, Lo and colleagues demonstrated that cell-free fetal DNA can be detected in the plasma and serum of pregnant women. Thus, Lo and colleagues were able to detect Y chromosomal DNA sequences in the plasma and serum of women carrying male fetuses. No Y chromosomal signals were seen in women carrying female fetuses. With the use of real-time quantitative polymerase chain reaction (PCR), Lo and colleagues showed that fetal DNA can be detected in maternal plasma during the first trimester and at a mean fractional concentration of approximately 3%. That figure is much higher than the fractional concentration of fetal nucleated cells in maternal blood. The high fractional concentration of fetal DNA in maternal plasma suggests that fetal DNA genotyping using this approach should be more readily achievable than previous methods based on the isolation of the rare fetal nucleated cells in maternal blood.
Another advantage of using plasma DNA is that fetal DNA is cleared rapidly from maternal plasma after delivery, with a half-life of approximately 16 minutes. This observation indicates that there is no risk of fetal DNA persistence from one pregnancy to the next, unlike the observation that certain fetal cell populations can persist for long periods within the mother’s body. In other words, fetal genotype information that could be obtained from maternal plasma DNA analysis reflects that of the current pregnancy.
It is now accepted that the placenta is the principal origin of such fetal DNA in maternal plasma. This conclusion is based on several observations. First, fetal DNA in maternal plasma can be detected at normal concentrations in anembryonic pregnancies in which only the placenta, but not other fetal tissues, is present. Second, fetal DNA in maternal plasma carries the DNA methylation signature of the placenta. Third, in cases in which the placenta carries a distinctive cytogenetic signature when compared with other fetal tissues, such a signature can also be seen in maternal plasma.
Prenatal diagnostic applications
The first diagnostic applications using fetal DNA in maternal plasma have focused on the detection of sequences that a fetus has inherited from the father but which are absent in the genome of the mother. Examples of such sequences involve those that are present on the Y chromosome. Several groups have reported the detection of fetal-derived Y chromosomal sequences in maternal plasma for prenatal gender determination for sex-linked diseases, such as hemophilia. Another application of prenatal gender determination using this approach is in congenital adrenal hyperplasia in which prenatal dexamethasone treatment for the prevention of virilization is only indicated for female fetuses.
Another early diagnostic application of fetal DNA in maternal plasma is for the prenatal determination of fetal RhD status in RhD-negative pregnant women. This application has been validated by several large-scale studies.
Due to the robustness with which fetal gender and RhD status can be determined from maternal plasma, these applications have been used diagnostically in several centers, initially in Europe and more recently in the United States.
Application to the hemoglobinopathies
Fetal DNA in maternal plasma was first applied to the hemoglobinopathies for the prenatal diagnosis of β-thalassemia. Chiu and colleagues developed a real-time PCR assay to detect the 4–base pair (bp) deletion in codons 41/42 of the human β-globin gene ( HBB ), which is the most common β-thalassemia mutation in China. They studied families in which the father was a carrier of this mutation whereas the mother was a carrier of another HBB mutation. Thus, the primary use of this approach is in the exclusion of β-thalassemia in the fetus, in which case the paternal mutation is not detected in maternal plasma. Provided that the assay has been optimized to have a high sensitivity for the detection of circulating fetal DNA, a negative signal indicates that no further prenatal testing is needed for a particular case. Because the diagnostic significance of this lack of detection of the paternal mutation is great, several investigators in the field have proposed developing a positive control for the detection of fetal DNA in maternal plasma (discussed later).
Conversely, if the paternal mutation is detected in maternal plasma, the only conclusion is that the fetus has inherited the paternal mutation. It is unclear, however, if a fetus is only a carrier or also has inherited the maternal mutation. This is a more challenging issue because the maternal mutation is present in maternal plasma after the release by the mother’s own cells, irrespective of fetal mutational status. Recent advances in highly quantitative digital counting technologies, such as digital PCR, have allowed the solution of this problem (discussed later).
Several investigators have studied the detection other paternally inherited HBB mutations in maternal plasma. Unlike the 4-bp deletion (described previously), many of these other mutations are caused by point mutations. Hence, the specificity of the detection method, with regard to the target sequence, needs to be high. One approach with which this can be done is using variants of allele-specific or single-allele extension reactions followed by mass spectrometry. This has been successfully implemented for the IVS2 654 (C →T), nt -28 (A → G), and CD 17 (A → T) mutations causing β-thalassemia and the (GAG→AAG) missense mutation in codon 26 of the HBB gene causing hemoglobin E disease. Furthermore, it has been shown that through the detection of a paternally inherited single nucleotide polymorphism (SNP) allele that is linked to the nonmutant HBB gene of the father, β-thalassemia major can be excluded. Several groups have demonstrated that arrayed primer extension assays can be used for the detection of paternally inherited fetal β-thalassemia mutations and linked SNPs in maternal plasma. Another promising approach is the use of single-molecule techniques, such as digital PCR (discussed later).
Another strategy that has been attempted by investigators is developing technologies that allow the selective enrichment of fetal DNA in maternal plasma. For this approach to work, physical or biochemical characteristics that could differentiate fetal from maternal DNA in maternal plasma have to be found. One characteristic is the size of the DNA molecules, because Chan and colleagues first reported that fetal DNA molecules in maternal plasma are shorter than their maternally derived counterparts. Li and colleagues were able to obtain a relative enrichment of the circulating fetal DNA by the use of a gel-based electrophoretic system and to selectively extract the short DNA molecules from the gel. Using this approach, Li and colleagues performed noninvasive prenatal detection of four HBB mutations in maternal plasma. This gel-based system is complex, however, and potentially prone to contamination to be used in a routine diagnostic context. Thus, it is hoped that future developments of more rapid and efficient size separation systems will allow this strategy to be used on a routine basis.
Recently, a proof-of-concept report has shown that fetal DNA in maternal plasma can be used for the noninvasive prenatal exclusion of hemoglobin Bart’s disease. The sensitivity reported in this preliminary report, however, which is based on microsatellite polymorphisms, is only 33.3%. Because an established ultrasound-based approach is already used for this purpose, it is currently unclear if the DNA-based approach offers any additional advantage over the more conventional ultrasound-based method.
Application to the hemoglobinopathies
Fetal DNA in maternal plasma was first applied to the hemoglobinopathies for the prenatal diagnosis of β-thalassemia. Chiu and colleagues developed a real-time PCR assay to detect the 4–base pair (bp) deletion in codons 41/42 of the human β-globin gene ( HBB ), which is the most common β-thalassemia mutation in China. They studied families in which the father was a carrier of this mutation whereas the mother was a carrier of another HBB mutation. Thus, the primary use of this approach is in the exclusion of β-thalassemia in the fetus, in which case the paternal mutation is not detected in maternal plasma. Provided that the assay has been optimized to have a high sensitivity for the detection of circulating fetal DNA, a negative signal indicates that no further prenatal testing is needed for a particular case. Because the diagnostic significance of this lack of detection of the paternal mutation is great, several investigators in the field have proposed developing a positive control for the detection of fetal DNA in maternal plasma (discussed later).
Conversely, if the paternal mutation is detected in maternal plasma, the only conclusion is that the fetus has inherited the paternal mutation. It is unclear, however, if a fetus is only a carrier or also has inherited the maternal mutation. This is a more challenging issue because the maternal mutation is present in maternal plasma after the release by the mother’s own cells, irrespective of fetal mutational status. Recent advances in highly quantitative digital counting technologies, such as digital PCR, have allowed the solution of this problem (discussed later).
Several investigators have studied the detection other paternally inherited HBB mutations in maternal plasma. Unlike the 4-bp deletion (described previously), many of these other mutations are caused by point mutations. Hence, the specificity of the detection method, with regard to the target sequence, needs to be high. One approach with which this can be done is using variants of allele-specific or single-allele extension reactions followed by mass spectrometry. This has been successfully implemented for the IVS2 654 (C →T), nt -28 (A → G), and CD 17 (A → T) mutations causing β-thalassemia and the (GAG→AAG) missense mutation in codon 26 of the HBB gene causing hemoglobin E disease. Furthermore, it has been shown that through the detection of a paternally inherited single nucleotide polymorphism (SNP) allele that is linked to the nonmutant HBB gene of the father, β-thalassemia major can be excluded. Several groups have demonstrated that arrayed primer extension assays can be used for the detection of paternally inherited fetal β-thalassemia mutations and linked SNPs in maternal plasma. Another promising approach is the use of single-molecule techniques, such as digital PCR (discussed later).
Another strategy that has been attempted by investigators is developing technologies that allow the selective enrichment of fetal DNA in maternal plasma. For this approach to work, physical or biochemical characteristics that could differentiate fetal from maternal DNA in maternal plasma have to be found. One characteristic is the size of the DNA molecules, because Chan and colleagues first reported that fetal DNA molecules in maternal plasma are shorter than their maternally derived counterparts. Li and colleagues were able to obtain a relative enrichment of the circulating fetal DNA by the use of a gel-based electrophoretic system and to selectively extract the short DNA molecules from the gel. Using this approach, Li and colleagues performed noninvasive prenatal detection of four HBB mutations in maternal plasma. This gel-based system is complex, however, and potentially prone to contamination to be used in a routine diagnostic context. Thus, it is hoped that future developments of more rapid and efficient size separation systems will allow this strategy to be used on a routine basis.
Recently, a proof-of-concept report has shown that fetal DNA in maternal plasma can be used for the noninvasive prenatal exclusion of hemoglobin Bart’s disease. The sensitivity reported in this preliminary report, however, which is based on microsatellite polymorphisms, is only 33.3%. Because an established ultrasound-based approach is already used for this purpose, it is currently unclear if the DNA-based approach offers any additional advantage over the more conventional ultrasound-based method.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree