Molecular Anatomy of the BCR-ABL1 Junction
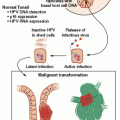
The breakpoints within ABL1 occur upstream of exon 1b, downstream of exon 1a, or more frequently, between the two. Regardless of the exact breakpoint location, splicing of the primary transcript yields an mRNA in which BCR sequences are fused to ABL1 exon a2. Breakpoints within BCR localize to one of three breakpoint cluster regions. More than 90% of CML patients and one-third of Ph+ ALL patients express the 210-kDa isoform of BCR-ABL1, in which the break occurs in the 5.8-kb major breakpoint cluster region (M-bcr), which spans exons e12-e16 (formerly exons b1-b5). Alternative splicing gives rise to either b2a2 (e13a2) or b3a2 (e14a2) transcripts,4 which are mutually exclusive and present in 36% and 64% of patients, respectively. Patients with b3a2 rearrangements are, on average, older than patients with b2a2 transcripts and have elevated platelet levels.5 The remainder of Ph+ ALL patients and rare CML cases harbor breakpoints further upstream in the 54.4-kb minor breakpoint cluster region (m-bcr), generating an e1a2 transcript that is translated into p190BCR-ABL1.6 A third breakpoint downstream of exon 19 in the micro breakpoint cluster region (μ-bcr) gives rise to an e19a2 BCR-ABL1 mRNA and p230BCR-ABL1 and is associated with neutrophilia. The reciprocal ABL1-BCR transcript, although detectable in approximately two-thirds of patients, does not seem to play any significant role in pathogenesis.7
Functional Domains of BCR-ABL1 and Kinase Activation
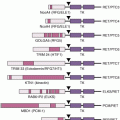
p210BCR-ABL1 contains several distinct domains (Fig. 26.1B).8 The N-terminal coiled-coil domain of BCR allows BCR-ABL1 dimerization, which is critical for kinase activation. The p210BCR-ABL1 protein also retains the serine/threonine kinase and Rho guanine nucleotide exchange factor homology (Rho-GEF) domains of BCR, which are deleted in p190BCR-ABL1, which may explain differences in disease phenotype associated with the two variants. In contrast to BCR, the ABL1 sequence is almost completely retained, including SRC homology domains 2 and 3, the tyrosine–kinase domain, a proline-rich sequence, and a large C terminus with nuclear localization signal, DNA-binding, and actin-binding domains. The N-terminal “cap” region of ABL1, which is lost in the BCR-ABL1 fusion, negatively regulates kinase activity by binding to a hydrophobic pocket at the base of the kinase domain, which, in the 1b isoform, is mediated by N-terminal myristoylation.
Signal Transduction
Numerous substrates and binding partners of BCR-ABL1 have been identified (see Fig. 26.1B) that contribute to increased proliferation, decreased apoptosis, defective adhesion to bone marrow stroma, and genetic instability.9 Because a comprehensive review of the multiple implicated pathways is beyond the scope of this chapter, we will focus on those for which strong evidence supports a rate-limiting role in disease pathogenesis.
Phosphatidylinositol-3 Kinase
Phosphatidylinositol-3 kinase (PI3K) is activated by autophosphorylation of tyrosine 177, which generates a high-affinity docking site for the GRB2 adapter, which in turn recruits GAB2 into a complex that activates PI3K. Consistent with a critical role of the Y177/GRB2/GAB2 axis, a mutation of this critical tyrosine to phenylalanine or a lack of GAB2 abrogates myeloid leukemia.10 An alternative pathway of PI3K activation is a complex formation between its p85 regulatory subunit, CBL, and CrkL, which bind to the SH2 and proline-rich domains of BCR-ABL1.11 PI3K activates the serine/threonine kinase AKT, which suppresses the activity of the forkhead O transcription factors (FOXO), thereby promoting survival.12 Additionally, PI3K enhances cell proliferation by promoting proteasomal degradation of p27 through upregulation of S-phase kinase-associated protein 2 (SKP2), the F-Box recognition protein of the SCFSKP2 E3 ubiquitin ligase.13 Another important outlet of PI3K signaling is the AKT-dependent activation of mammalian target of rapamycin (mTOR), which enhances protein translation and cell proliferation.14
Rat Sarcoma/Mitogen-Activated Protein Kinase Pathways
GRB2-mediated recruitment and activation of son of sevenless (SOS) promotes exchange of guanosine triphosphate (GTP) for GDP on rat sarcoma (RAS).15 GTP-RAS activates mitogen-activated protein kinase (MAPK), promoting proliferation. Signaling from RAS to MAPK involves the serine/threonine kinase RAF-116 and ras-related C3 botulinum toxin substrate (RAC), another GTP–GDP exchange factor.17 A crucial role for the latter is supported by the fact that a lack of RAC1/2 delays BCR-ABL1–driven leukemia in a murine model.
Janus Kinase/Signal Transducer and Activator of Transcription Pathway
BCR-ABL1 activates signal transducer and activator of transcription 5 (STAT5) through direct phosphorylation or indirectly through phosphorylation by hematopoietic cell kinase (HCK), a SRC family kinase, or janus kinase 2 (JAK2).18 Active STAT5 induces the transcription of antiapoptotic proteins like myeloid cell leukemia 1 (MCL-1) and B-cell lymphoma-extra large (Bcl-xL).19 JAK2 has been shown to play a central role in the cytokine signaling machinery that enables survival of CML stem cells in the presence of BCR-ABL1 tyrosine kinase inhibitors. Recent studies have also shown that the complete lack of STAT5 abrogates both myeloid and lymphoid leukemogenesis, implicating it as a potential target for the elimination leukemic stem cells.20–22
Cytoskeletal Proteins
BCR-ABL1 phosphorylates several proteins involved in adhesion and migration, including focal adhesion kinase (FAK), paxillin, p130 CrK-associated substrate (p130CAS), and human enhancer of filamentation 1 (HEF1). This and the activation of RAS23 are thought to impair integrin-mediated adhesion of CML progenitors to stroma and the extracellular matrix, causing premature circulation as well as abnormal proliferation of Ph+ progenitors.24 A novel adaptor protein pathway in which BCR-ABL1 interaction with Grb2-related adaptor downstream of Shc (GADS)/SH2 domain containing leukocyte protein of 76kDa (SLP-76)/non-catalytic region of tyrosine kinase adaptor protein 1 (NCK1) regulates the actin cytoskeleton and nonapoptotic membrane blebbing was also recently described.25
DNA Repair
BCR-ABL1 impairs DNA damage surveillance by various mechanisms. For example, BCR-ABL1 has been shown to suppress checkpoint kinase 1 (CHK1) through the inhibition of ataxia telangiectasia and Rad3-related protein (ATR)26 or the downregulation of breast cancer 1, early onset (BRCA1), a substrate of ataxia telangiectasia mutated (ATM).27 Nonhomologous end joining and homologous recombination, both critical double-strand break repair pathways, are defective in CML. BCR-ABL1 also upregulates RAD51, inducing rapid but low-fidelity double-strand break repairs on challenge with cytotoxic agents and inducing reactive oxygen species (ROS) that promote chronic oxidative DNA damage, double-strand breaks, and point mutations. It has been demonstrated that BCR-ABL1 kinase inhibits the activity of uracil-DNA glycosylase (UNG2), which leads to the accumulation of uracil derivatives in genomic DNA and contributes to increased point mutations.28 Lastly, telomere length decreases with disease progression from CP to BP.29
Although significant progress has been made to understand the extraordinary complexity of CML biology, a complete picture is still elusive. To overcome the limitations of investigating single pathways, quantitative proteomics30 and whole transcriptome analyses31 are being used to establish a comprehensive picture of BCR-ABL1 signaling. These results suggest that cellular processes in CML, rather than relying on a single pathway, use integrated networks to fully realize their leukemogenic potential.
Murine Models of Chronic Myeloid Leukemia
The most commonly used murine model of CML is the retroviral expression of BCR-ABL1 in bone marrow followed by transplantation into lethally irradiated syngeneic recipients, which develop a CML-like myeloproliferative neoplasm.32 Recently, an inducible transgenic mouse model has been developed, in which conditional BCR-ABL1 expression is under the control of the three enhancers of the murine stem cell leukemia (SCL) gene. This model serves as a promising new tool for studying leukemogenic mechanisms in hematopoietic stem cells during disease initiation and progression.33 Lastly, xenograft models use various strains of immunodeficient mice for engraftment of primary CML cells.34 A limitation of xenograft models is low engraftment of CML CP cells, probably because of their compromised interactions with the microenvironment as a result of species differences in cytokines and adhesion molecules. Promising results have been obtained by injecting CML cells directly into the livers of newborn mice.35
Chronic Myeloid Leukemia Stem Cells
The origin of CML in a pluripotent hematopoietic stem cell (HSC) was elegantly demonstrated in the late 1970s.36 BCR-ABL1 does not confer self-renewal, implying that it must be acquired by an HSC already endowed with this capacity.37 For unknown reasons, the main cellular expansion occurs in the progenitor cell compartment, while, at least initially, the majority of HSCs are Ph negative.38 Serial xenograft studies have shown that CML leukemia stem cells (LSC) reside within the quiescent CD34+38− fraction of bone marrow cells. Significant progress has recently been made by the identification of the interleukin 1 (IL-1) receptor–associated protein (IL-1RAP) as a surface marker specifically expressed on CD34+38− CML LSC.39 Several genes were shown to have a critical role for LSC maintenance in CML, including promyelocytic leukemia (PML),40 Rac2 GTPase,41 smoothened (SMO)/hedgehog (Hh),42 Wnt/β-catenin,43,44 phosphatase and tensin homolog (PTEN),45 hypoxia-inducible factor 1 (HIF-1),46 B lymphoid kinase (BLK),47 stearoyl-CoA desaturase1 (SCD1),48 transforming growth factor beta (TGF-β), and FOXO3a.12 Additionally, BCL6 was reported to be required for the maintenance of LSCs in CML and contribute to drug resistance.49 BCL6 expression in these cells is regulated in a PTEN/AKT/FOXO-dependent manner. Recently, an interesting new role of lipid metabolism has emerged in CML stem cell maintenance due to the increased expression of arachidonate 5-lipoxygenase (ALOX5). Alox5 knockout mice fail to develop CML, suggesting a critical role of ALOX5 in CML leukemogenesis.50 Sirtuin 1 (SIRT1), a nicotinamide adenine dinucleotide-dependent protein deacetylase that promotes cell survival under metabolic, oxidative, and genotoxic stresses through deacetylation of multiple substrates including p53, Ku70, and FOXO, is also transcriptionally activated by BCR-ABL1. SIRT1 knockdown or inhibition by a small-molecule inhibitor effectively suppresses the development of CML-like myeloproliferative disease in mice.51 Importantly, it has been shown that CML stem cell survival may be independent of BCR-ABL1 kinase activity.52 A number of studies further suggest that the bone marrow microenvironment provides survival signals to LSCs by involving a number of mechanisms such as chemokine (C-X-C motif) receptor 4 (CXCR4)/stromal cell-derived factor 1 (SDF-1),53,54 N-cadherin, and Wnt/β-catenin.55 However, the fact that many of these genes are also critical for maintenance and self-renewal of normal HSCs may be an obstacle to exploiting them as therapeutic targets.
Progression to Blastic Phase
Disease progression is believed to be due to the accumulation of molecular abnormalities that lead to a loss of terminal differentiation capacity of the leukemic clone, which continues to depend on BCR-ABL1 activity. BCR-ABL1 mRNA and protein levels are higher in CML-BP than in CP cells, including CD34+ granulocyte macrophage progenitors (GMP), which are expanded in BP.56 One of the mechanisms that enhances BCR-ABL1 activity in BP is inactivation of the phosphatase protein phosphatase 2A (PP2A) through upregulation of SET.57,58 Constitutive BCR-ABL1 activity has also been shown to perturb the CML transcriptome,59 resulting in altered expression of genes implicated in BP (e.g., preferentially expressed antigen in melanoma [PRAME], myeloid zinc finger 1 [MZF1], ecotropic virus integration site 1 [EVI-1], Wilms tumor 1 [WT1], and JUN-B). Interestingly, a six-gene signature (NIN1/RPN12 binding protein 1 homolog [S. cerevisiae] [NOB1], DEAD [Asp-Glu-Ala-Asp] box polypeptide 47 [DDX47], immunoglobulin superfamily member 2 [IGSF2], lymphotoxin beta receptor 4 [LTBR4], scavenger receptor class B, member 1 [SCARB1], and solute carrier family 25 member A [SLC25A3]) was recently found to accurately discriminate early from late CP, CP from AP, and CP from BP60; however, the biologic role of these genes in disease progression is still unknown.
CML-BP patients also harbor various additional genetic lesions such as additional chromosomes, gene insertions and deletions, and/or point mutations. A deep-sequencing study of a small cohort of CML-BP patients detected mutations in 76.9% of cases.61 The most common mutations (other than those in the BCR-ABL1 kinase domain) occur at the loci of the runt-related transcription factor (RUNX1),62 the additional sex combs like 1 (ASXL1), WT1, and the tumor suppressor gene TP5363 in myeloid BP and in cyclin-dependent kinase inhibitor 2A/2B (CDKN2A/B), and the Ikaros transcription factor (IKZF1) in lymphoid BP.64
The most striking feature of BP, the loss of differentiation capacity, suggests that the function of key myeloid transcription factors must be compromised. Occasionally, the differentiation block can be ascribed to mutations that result in the formation of dominant-negative transcription factors such as runt-related transcription factor 1- ecotropic virus integration site 1 (AML1-EVI-1) or nucleoporin 98kDa-homeobox A9 (NUP98-HOXA9), which block differentiation or favor preferential growth of immature precursors.65,66 Isolated cases of myeloid transformation have been associated with the acquisition of core binding factor mutations typical of AML. A more universal mechanism appears to be the BCR-ABL1–induced downregulation of CCAAT/enhancer binding protein-α (CEBPα) through the stabilization of the translational regulator heterogeneous nuclear ribonucleoprotein E2 (hnRNP E2), which is low or undetectable in CP but readily detectable in CML-BP.67
Aberrant Wnt/β-catenin activation cooperates with interferon-regulatory factor 8 (Irf8)68 to contribute to CML progression by conferring self-renewal capacity to GMPs.69 The acquisition of self-renewal by GMPs is expected to greatly increase the pool of LSCs in BP. Recently, a crucial role of the RNA-binding protein Musashi2 (MSI2) was shown in CML progression to BP, where MSI2 represses the expression of Numb, a protein that impairs the development and propagation of BP.70 Interestingly, expression microarray studies have implicated a few genes such as β-catenin not only in disease progression, but also in resistance to tyrosine–kinase inhibitors, supporting the view that drug resistance and disease progression share a common genetic basis.71 This has implications for prognostication as well as for the development of strategies to prevent progression and overcome resistance.
Conclusions
BCR-ABL1 orchestrates an integrated network of signaling pathways that upend the physiologic control of proliferation, cell death, DNA repair, and microenvironment interaction and lead to the clinical phenotype of CML. Cooperation with additional genetic events that accumulate over time inevitably leads to BP and drug resistance. Although significant progress has been made toward understanding transformation and disease progression, much remains to be learned. Efforts toward determining the molecular pathways critical for the maintenance of these cells, and to develop better and faster techniques to differentiate the LSC from the normal HSC, have been intensified in the last decade. The availability of genomewide scanning tools has undoubtedly accelerated this process. This knowledge has been used to design new strategies to target LSCs and hopefully lead to the discovery of new therapeutic targets to eliminate CML stem cells, overcome drug resistance unless effective therapy is initiated early on, and improve the prognosis of patients whose disease has progressed on therapy.
CHRONIC LYMPHOCYTIC LEUKEMIA
CLL is one of the most common leukemias in adults and has a relatively consistent immunophenotype, including dim surface immunoglobulin expression, CD19, CD20, CD23, along with the pan T-cell marker CD5.72 The impact on overall survival in both young and elderly patients with CLL is substantial: patients diagnosed under the age of 50 have a median expected life span of 12.3 years, compared to 31.2 years in an age-matched control group.73 Although younger patients have poor outcomes and shortened survival with CLL, several studies have also identified elderly patients as a high-risk group for poor survival following treatment.74–77 A subset of CLL patients have indolent disease for many years and do not require therapy. Improving our understanding of the origin, biology, and progression of CLL will improve risk stratification and will help identify new treatments for this disease.
Origin of Chronic Lymphocytic Leukemia
The identification of a normal B-cell counterpart remains controversial.78–80 Unlike most other B-cell lymphomas and leukemias (with the notable exception of mantle cell lymphoma), CLL coexpresses typical mature B-cell markers with CD5. This prompted many to hypothesize that CLL may be derived from CD5+ B cells whose immunoglobulin (Ig) VH is unmutated. However, the overall phenotype of CLL with expression of CD5, CD23, and CD19, and low levels of surface IgM or IgD is not observed in any normal B-cell counterpart. Additionally, investigators identified that approximately 40% of CLL cases have an unmutated IGHV locus, whereas the remainder are mutated.81,82 These two groups were also shown to have distinct clinical features, prompting the hypothesis that CLL may represent two distinct diseases.81,82
In contrast, two seminal articles examining gene expression profiling in CLL and normal B cells provided findings suggesting that CLL is in fact one disease with a common CLL gene signature.83,84 The first, by Klein et al.,83 examined mRNA profiles derived from IGHV unmutated, IGHV mutated, and normal B cells from different stages of differentiation. An unsupervised analysis of gene expression profiles demonstrated that IGHV mutated and unmutated CLL cases were not distinguished in any manner among a common profile typical of CLL. This CLL profile in the majority of samples most resembled postgerminal center memory B cells and lacked any similarity to naïve B cells, CD5+ B cells, or germinal center centroblasts. A supervised analysis of IGHV-unmutated and mutated CLL did, however, demonstrate distinct genes that could separate these two clinical subsets of CLL.
A second article, published concurrently by Rosenwald et al.,84 demonstrated similar findings of a common CLL profile described by Klein et al., as compared with other normal B cells and B-cell malignancies. In particular, the CLL gene phenotype was not shared by CD5+ normal B cells, thereby providing corroborating evidence that this is likely not the CLL cell of origin. In an unsupervised analysis of CLL samples, IGHV-unmutated and mutated samples were intermingled. However, a supervised analysis of IGHV-unmutated and mutated CLL again identified a number of genes differentially expressed in the former group related to B-cell receptor signaling and proliferation. In particular, ZAP70 (zeta-chain associated protein kinase) was overexpressed in IGHV-unmutated CLL as compared with IGHV-mutated CLL.85–88
Subsequent studies have suggested that ZAP-70 expression may partly explain why IGHV-unmutated CLL patients show more B-cell receptor signaling activity upon ligation of the B-cell receptor.89–91 Multiple studies confirming both the clinical prognostic significance of IGHV mutational status and/or ZAP-70 expression have subsequently been reported. IGHV status and/or ZAP70 represent very strong independent variables in predicting early disease progression, treatment remission duration, and survival of CLL patients. The variability in direct measurement of ZAP-70 among investigators has limited the application of this biomarker clinically, but more recently, ZAP70 methylation status has been demonstrated to be a clinically relevant surrogate.92 In the future, the assessment of methylation may supplant the direct measurement of ZAP-70.
Chromosomal Abnormalities in the Pathogenesis of Chronic Lymphocytic Leukemia
In CLL, conventional metaphase cytogenetics can identify chromosomal aberrations in only 20% to 50% of cases because of the low in vitro mitotic activity of CLL tumor cells.93 Early unstimulated metaphase karyotype studies of CLL demonstrated abnormalities, including trisomy 12, deletions at 13q14, structural aberrations of 14q32, and deletions of 11q, 17p, and 6q in descending frequency of occurrence.94
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree