Fig. 5.1
Molecular pathogenesis of colorectal dysplasia and cancer in inflammatory bowel disease. Summary of the molecular signals, mediators, gene targets, and mechanisms implicated in the progression from mucosal inflammation to dysplasia to cancer. Abbreviations: AID activation-induced cytidine deaminase, IL interleukin, TLR Toll-like receptor, TNF tumor necrosis factor, TSG tumor suppressor gene
5.2 Inflammation
During inflammation, the destiny of an epithelial cell is determined by the balance between pro- and anti-tumorigenic immune responses. Inflammation participates in the three main stages of carcinogenesis: tumor initiation, tumor promotion, and tumor progression. Tumor initiation defines the process by which a normal cell becomes premalignant. The inflammatory condition, in which the levels of cytokines, chemokines, and reactive oxygen and nitrogen species are increased, induces DNA mutations, epigenetic alterations, and genomic instability, all of which contribute to tumor initiation [1, 2]. Tumor promotion leads to the proliferation of genetically altered cells, which is enhanced by inflammation via the acceleration of anti-apoptosis, proliferation, and angiogenesis [1, 2]. Finally, genetic changes influenced by inflammation advance tumor spread from the primary site to multiple distant sites (metastasis) [1, 2]. Thus, inflammation and malignant tumor formation are closely connected at all stage of tumorigenesis.
Much of the current evidence regarding the inflammatory mediators of CAC comes from murine models, which have provided insights into the carcinogenic process [3]. For example, IL-10-deficient mice develop spontaneous colitis and colonic neoplasms similar to those that occur in patients with CD [4]. Mice can also be treated with the mucosal irritant dextran sulfate sodium (DSS), with or without the mutagenic agent azoxymethane (AOM), which induces damage similar to that seen in UC patients, following a dose-repeated oral administration, colitis, and colonic neoplasm [5–7]. Through animal models, it is now known that inflammatory cytokines, chemokines, cell-surface receptors, and microbiota in the gut play crucial roles in colitis-associated carcinogenesis.
Cytokines exhibit both pro-inflammatory and anti-inflammatory effects, the balance of which plays an important role in CAC [3]. TNF-α, a pro-inflammatory cytokine, is an important mediator of chronic inflammatory disease, including IBD. TNF-α levels are consistently upregulated in the blood and colonic mucosa of patients with IBD [8]. In carcinogenesis, TNF-α acts as a tumor initiator by stimulating the production of reactive oxygen species, which cause oxidative stress and thus DNA damage and mutations and, in turn, tumor promotion, by altering cell proliferation and cell death [9–11]. Consistent with the critical role of TNF-α in chronic inflammation, anti-TNF-α monoclonal antibody biologics, such as infliximab, adalimumab, and certolizumab pegol, have proven to be effective in the treatment of IBD [10]. Furthermore, in an in vivo study using a mouse model of colitis, a monoclonal anti-TNF-α antibody reduced the development of tumors [2]. These results confirm the critical role of TNF-α in chronic inflammation and CAC. The pro-inflammatory cytokine IL-6 mediates a wide variety of inflammation-associated diseases. IL-6 levels are increased in IBD, CRC, and CAC and contribute to CAC development by promoting the survival of neoplastic epithelial cells in the colon [12–15]. Thus, the blockage of IL-6 signaling transduction could be a useful therapeutic system for the treatment of CAC [16]. IL-6 is also involved in promoting the growth and tumorigenesis of colon cancer cells by altering the epigenome, such as silencing of DNA methyltransferase 1 (DNMT1)-mediated tumor suppressor genes [17]. Therefore, IL-6 appears to play a critical role in inflammation-associated carcinogenesis, although the molecular mechanisms are still being elucidated. Another important regulator of the immune response is the anti-inflammatory cytokine IL-10; its function is to block nuclear factor-kappaB (NF-kB) activity and to regulate the Jak-Stat signaling pathway [18, 19]. In addition, IL-10 downregulates TNF-α, vascular endothelial growth factor, and IL-6 production, which may account for its inhibitory effect on the tumor stroma [20]. IL-10 null mice develop spontaneous, generalized colitis and CAC in the presence of enteric bacteria. [21], whereas the administration of IL-10 improves the colitis and reduces tumor development by 50 %, even after colitis establishment [22]. Therefore, an incorrect balance of both pro- and anti-inflammatory cytokines is critical to both inflammation and inflammation-associated carcinogenesis.
Intraluminal bacterial endotoxins, TNF-α, and other pro-inflammatory cytokines act through extracellular receptors such as Toll-like receptors (TLRs) to initiate the phosphorylation cascades that transmit signals to key transcription factors such as NF-kB [23, 24]. TLR-4, which specifically responds to bacterial lipopolysaccharide ligand and is expressed at low levels in normal intestinal mucosa, is upregulated in the mucosa of patients with IBD, in the ileal mucosa of UC patients with pouchitis, in UC-associated CRC, and in colon tumors that develop in the AOM/DSS mouse model [25–27]. By contrast, TLR-4-deficient mice develop fewer and smaller tumors and produce less Cox-2 and prostaglandin E2, both of which mediate colorectal tumorigenesis. Conversely, mice deficient in the cytoplasmic immune receptor Nod1 and treated with AOM/DSS develop more severe colitis and larger tumors in the colonic mucosa compared to controls [28]. Taken together, these observations show that interactions between commensal bacterial components, elements of the innate immune response, and inflammation-induced tumorigenesis [29] are important for the initiation and maintenance of both chronic inflammation and tumor progression, through processes that involve NF-kB-regulated cytokines, chemokines, angiogenic factors, anti-apoptotic factors, and matrix proteases.
The mechanisms by which inflammatory cytokines promote the epithelial DNA mutations necessary for the initiation of neoplasia remain largely undefined. However, a potential mechanism has recently emerged from the study of activation-induced cytidine deaminase (AID), an enzyme that under physiologic conditions regulates class switching and somatic hypermutation in the immunoglobulin genes of activated B cells. AID induction by pro-inflammatory cytokines in human colon cancer cell lines was shown to lead to the accumulation of mutations in the tumor suppressor gene p53 [30]. AID is overexpressed in both neoplastic and nonneoplastic colonic epithelium from patients with IBD, as well as in some sporadic colon cancers. Therefore, this mechanism may account for the production of other potentially carcinogenic mutations in colonic epithelial cells in response to the overexpression of pro-inflammatory cytokines.
5.3 Oxidative Stress
Inflammation gives rise to colonic carcinogenesis by generating oxidative stress. IBD has been viewed as an oxyradical overload disease, in which long-standing inflammation increases the risk of malignant tumors [31]. Oxidative stress causes cellular damage that contributes to the pathogenesis of the colitis itself and to colon carcinogenesis. The initiation of tumor formation in chronic inflammatory tissue might be induced by reactive oxygen and nitrogen species (RONS), which are released by cells of the innate immune system. The inflamed colonic mucosa of patients with active IBD-associated colitis is characterized by the increased expression of nitric oxide synthase (NOS) and RONS [32–34]. Additionally, McKenzie et al. demonstrated the oxidation of thiols in the active site of glyceraldehyde-3-phosphate dehydrogenase, with subsequent inhibition of enzyme activity, in colonic epithelial cells from the inflamed mucosa of patients with IBD but not from paired samples of non-inflamed mucosa [35]. Furthermore, measurements of 8-hydroxydeoxyguanosine in mucosal biopsies from patients with UC showed that oxidative DNA damage progressively accumulates with the increasing duration of UC, reaching maximal levels in dysplastic lesions. This observation has implications for mutagenic and carcinogenic progression.
There is also evidence regarding the mechanisms of colorectal carcinogenesis induced by oxidative stress. Free radicals affect many metabolic processes, including those that regulate DNA, RNA, proteins, and lipids [31, 36]. When free radicals alter the genes or proteins that maintain homeostasis in intestinal epithelial cells, for example, p53, a dysplastic lesion forms and ultimately CAC. Hussain and coworkers [37] compared mutations of p53 in biopsy samples collected from the inflamed colonic mucosa of UC patients and from the normal mucosa of individuals without UC. The majority of the UC samples had a high frequency of p53 mutations compared with the control. In addition, these mutations were found only in inflamed not in non-inflamed mucosa. Another group showed that hydrogen peroxide inactivated the mismatch repair (MMR) system in colorectal cancer cell lines, apparently by damaging the protein complexes responsible for DNA repair [38] and causing microsatellite instability (MSI). In fact, MSI has been detected in the chronic inflammatory mucosa of UC patients, even in those with short disease duration, before the risk of dysplasia or CAC increases [39]. By contrast, MSI was not found in the colonic mucosa of healthy controls or that of patients with benign colitis or that of patients with CD [39, 40]. Thus, only a specific type of UC-induced colitis appears to cause MSI.
Several animal models have demonstrated that RONS participate in colitis-induced carcinogenesis. The application of peroxynitrite to the rat rectum was shown to cause colonic inflammation [41]. Scavengers of oxygen radicals (such as superoxide dismutase), catalase, and NOS inhibitors attenuate inflammation of the colon in animal models of chemically induced colonic injury [42]. Likewise, mice with inducible knockout of NOS develop attenuated colitis in response to injury [43], and APCmin/+ mice, carrying a mutation in the adenomatous polyposis coli gene that causes multiple intestinal adenomas, developed fewer adenomas when they were crossed with mice carrying an inducible knockout of NOS or given an inducible NOS inhibitor. Although APCmin/+ mice are not considered to be models of colon cancers that arise from colitis, these data support the concept that oxidative stress promotes colon carcinogenesis. The fact that mice deficient in glutathione peroxidase enzymes develop inflammation and cancer of the small and large intestines also supports a model in which antioxidant pathways prevent the transition of inflammation to neoplasia.
5.4 Genetic Instability
Genetic instability can be divided into two clinically distinct forms, both of which have been extensively studied in CRC: chromosomal instability (CIN) and MSI [44]. CIN and MSI are detected with the same frequency in CAC (85 % CIN, 15 % MSI) as in sporadic CRC [45]. CIN is manifested by genomic alterations that affect large DNA segments, resulting in aneuploidy, translocations, deletions, gene copy amplifications, and telomere shortening. It is typically associated with the progressive accumulation of mutations in onco-suppressor genes (APC, p53) and oncogenes (KRAS). MSI can be preceded by the alternation/inactivation of DNA repair mechanisms, including nucleotide excision repair, base excision repair, and MMR [46]. Furthermore, besides the many genetic contributions to CIN and MSI, epigenetic elements can affect tumor initiation, proliferation, and metastasis. In particular, the hypermethylation of onco-suppressor DNA promoter regions and microRNAs are major epigenetic mechanisms of gene silencing in colorectal carcinogenesis [47].
5.4.1 Chromosomal Instability
The extent and types of CIN associated with IBD-related dysplasia and CAC have been evaluated by comparative genomic hybridization, fluorescent in situ hybridization (FISH), flow cytometry, and DNA fingerprinting. CIN in IBD, in addition to its similar frequency to CIN in sporadic carcinogenesis, also affects many of the same loci and results in similar mean numbers of chromosomal alterations per case [45, 48]. An important distinction, however, is that CIN in UC is distributed broadly, involving even mucosa that is negative for and remote from dysplasia, whereas in sporadic CRC it is restricted to tumor tissues [45, 49, 50]. Chromosomal alterations therefore appear to occur early in the course of IBD-related neoplastic progression and prior to the histologic features of dysplasia. CIN is typically absent in patients with UC who do not also harbor dysplasia or CAC. Thus, the presence or absence of CIN in nonneoplastic mucosa has been advanced as a marker enabling patients to be stratified into distinct categories of progressors and non-progressors, respectively [45, 49, 51–53]. Progressors have an earlier onset and longer duration of disease [54]. Recently, the FISH-based detection of combined alterations in four key chromosomes in nonneoplastic mucosa was reported to be 100 % sensitive and 92 % specific in distinguishing progressors from non-progressors, suggesting the application of this method to improve surveillance [55]. One proposed mechanism by which chronic inflammation could lead to CIN involves accelerated telomere shortening. The telomeres of colonic epithelial cells from patients with UC are shorter than those of the colonic epithelial cells of healthy controls. This difference is attributable to the faster cell turnover, increased replication, and increased oxidative damage that result from repeated cycles of injury and regeneration [56, 57]. Telomere shortening in turn correlates with CIN, as reflected by the higher rates of chromosomal arm and centromere loss and the higher frequency of anaphase bridges in the colonic epithelium of patients with UC who have dysplasia or CAC but not in that of controls [53]. Telomere shortening beyond a critical point is associated with aging as well as growth arrest, through replicative senescence when DNA damage checkpoints are intact, but through chromosomal damage, such as breaks and end-to-end fusions, when these checkpoints are defective [58, 59]. Thus, telomere shortening could facilitate CIN, provided that DNA damage checkpoints are somehow also inactivated through mutations of checkpoint genes such as p53. The rates of telomere shortening in the colonic epithelial cells of patients with UC are double those seen in healthy controls and occur mostly during the first 8 years of disease, the time frame when the risk of CRC begins to become clinically significant [57].
Aneuploidy occurs in approximately 33 % of patients with long-standing UC, in 20–50 % of dysplastic lesions, and in 50–90 % of cancers [60]. Regions of aneuploidy in the large bowel of UC patients are frequently those exhibiting dysplasia. Since aneuploidy precedes the appearance of histologic changes, it may be a useful marker of developing neoplastic lesions in UC patients. Despite the utility of flow cytometry in assessing aneuploidy in patients with IBD, it is not widely available and thus cannot be universally applied in the follow-up of patients with long-standing UC.
The timing and frequency of DNA mutation differs in CAC vs. sporadic CRC (Fig. 5.2). Loss of APC function, an early event in the progression of sporadic CRC, is less frequent and usually occurs at the late stage of CAC development [61–63]. APC mutations are rarely, if ever, detected in cells of the colitic mucosa that are negative or indefinite for dysplasia, and <14 % of tissues with low-grade dysplasia or CAC have mutations in APC [61, 62, 64]. Likewise, allelic deletion of APC occurs in <33 % of CAC cases [64]. Loss of p53 function is an important step in CAC progression, and allelic deletion of p53 is observed in 50–85 % of CACs [65, 66]. Loss of heterozygosity (LOH) at p53, which correlates with malignant progression, was detected in 6 % of biopsy samples without dysplasia, 9 % with indefinite dysplasia, 33 % with low-grade dysplasia, 63 % with high-grade dysplasia, and 85 % with CAC [65]. Mutations in p53 are found in the colon tissue of patients with colitis and often in mucosa that is nonneoplastic or only indefinite for dysplasia [65–67]. In carefully mapped colectomy specimens, p53 mutations were shown to occur early in tumorigenesis, before aneuploidy [66, 67]. In fact, mutations were found in inflamed mucosa from >50 % of UC patients who did not have CAC, indicating that chronic inflammation caused these mutations [37]. Mutations of the KRAS proto-oncogene, present in 40–60 % of sporadic CRCs, are probably an early event in these cancers [68], whereas they occur at a lower but significant frequency (24 %) in CAC [69]. Thus, KRAS mutations probably play a significant role in the later stage of CAC. The tumor suppressor gene Rb is often mutated or lost in epithelial tumors. In UC-associated carcinogenesis, Rb LOH is present in 20 % of dysplasia and 30 % of CAC specimens [70]. Finally, losses at chromosome 18q are relatively rare events in the sequence of dysplasia → carcinoma in UC. LOH of 18q, the site of the deleted in colorectal carcinoma gene, was observed in 12 % of CACs and 33 % of dysplasias but not in nonneoplastic lesions or inflamed mucosa [71].
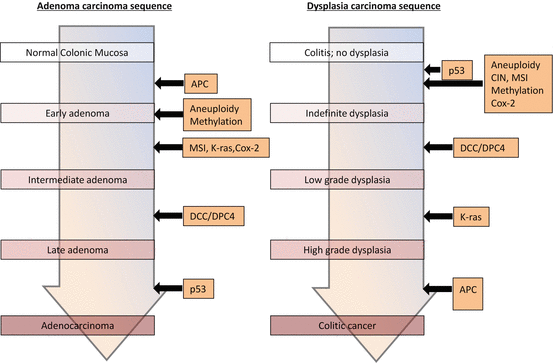

Fig. 5.2
Comparison of the molecular alterations in sporadic colon cancer (left) and colitis-associated colorectal cancer (right). There are similarities between the two pathways, including the development of chromosomal instability, microsatellite instability, DNA methylation, activation of the KRAS oncogene and of cyclo-oxygenase (Cox)-2, and mutation and the eventual loss of heterozygosity of p53, the adenomatous polyposis coli (APC) gene, and the deleted in colorectal carcinoma (DCC) genes DCC/DPC4. However, the frequency and sequence of these events differ between the pathways
5.4.2 Microsatellite Instability
Microsatellites are short repetitive sequences (one to five nucleotides) of DNA that are randomly distributed throughout the whole genome. MSI involves the loss of function of several genes (hMSH2, hMLH1, hPMS1, hPMS2, hMSH6, and hMLH3) that repair DNA base-pair mismatches. DNA MMR deficiency results in a strong mutator phenotype and MSI [72]. The stability of these sequences is a good measure of the general integrity of the genome. MSI reflects a gain or loss of repeat units in a germline microsatellite allele, consistent with the clonal expansion that is typical of cancer. UC-associated carcinogenesis can also be associated with MSI. A high rate of MSI in long-standing UC is probably related to the genomic instability produced by repeated inflammatory stimulation, and the influence of inflammation has been evaluated in estimates of MSI in UC [73]. Indeed, although the molecular mechanisms involved in the increased risk of CAC in UC are for the most part still unclear, many appear to be related to MSI [39]. The prevailing hypothesis is that the overproduction of free radicals overwhelms the ability of the cell to repair oxidative DNA damage prior to replication [39, 74]. Alternatively, prolonged and repeated oxidative insults may directly inactivate DNA MMR genes [38]. One study reported that half of UC mucosal samples with high MSI exhibit MLH1 hypermethylation [75]. However, in contrast to colon cancer in patients with hereditary non-polyposis, there is little evidence for MMR defects as a cause of MSI in UC [76, 77]. A recent report described the adaptive increased activity of 3-methyladenine DNA glycosylase (AAG) and apurinic endonuclease (APE1) in areas of the UC colon undergoing active inflammation [78]. This imbalanced increase appeared to be associated with the MSI characteristic of UC. These data were consistent with a possible novel mechanism by which the colonic cells of patients with chronic colonic inflammation acquire MSI. In UC patients, areas of the colonic epithelium with active inflammation exhibited increased AAG and APE1 enzyme activity; the largest increases and imbalances occurred in areas with inflammation as well as MSI. These observations suggest that the adaptive imbalanced increase in these enzymes has DNA-damaging effects and contributes to carcinogenesis in the chronically inflamed colon [78].
5.4.3 Aberrant DNA Methylation
Epigenetics, which includes histone modifications and DNA methylation, alters gene expression without changing the DNA sequence and can be transmitted to daughter cells. There is also significant cross talk between histone modifications and DNA methylation, both of which are highly dysregulated in many diseases, particularly in cancer [79, 80]. Epigenetic alterations are observed during inflammation and inflammation-associated carcinogenesis [81, 82]. DNA methylation involves the addition of a methyl group to the fifth carbon position in the pyrimidine ring of cytosine located in the context of CpG dinucleotides. It is one of the most well-studied epigenetic processes and is maintained during replication by the enzyme DNMT1, whereas de novo DNA methylation is believed to be mediated by DNMT3A and DNMT3B. However, there is functional overlap between DNMT1, DNMT3A, and DNMT3B [83–85].
DNA methylation contributes to the development and progression of CAC. Methylation of CpG islands in several genes precedes dysplasia and can be detected throughout the mucosa of patients with UC [86]. Among CAC tissues from UC patients, hMLH1 hypermethylation was recognized in six of 13 specimens with high levels of MSI, one of six with low levels of MSI, and four of 27 without MSI [75], suggesting that methylation induces MSI. The attenuated expression of the cell cycle inhibitor gene p16INK4a is associated with sporadic CRC; p16INK4a is also frequently hypermethylated in neoplastic tissues from UC patients. Approximately 10 % of nonneoplastic lesions have hypermethylation of the p16 promoter; the rate increases with higher grades of dysplasia, reaching 100 % in CAC [87]. Additionally, p14ARF, an indirect regulator of p53, is encoded by the same gene as p16INK4a. Loss of expression of p14ARF by hypermethylation is frequently observed in the mucosa of UC patients. In one study, it was detected in 50 % of CAC, 33 % of dysplastic, and 60 % of nonneoplastic mucosal samples collected from UC patients compared with 3.7 % of normal colonic mucosa samples [88]. Another group investigated the methylation status of ten genes [p16, p14, runt-related transcript factor-3 (RUNX3), Cox-2, E-cadherin, methylated-in-tumor-1 (MINT1), MINT31, HPP1, estrogen receptor, and SLC5A8] in tissue samples from CAC and in nonneoplastic colonic mucosa from UC patients with and without neoplasia [89]. Methylation of the promoters of RUNX3, MINT1, and COX-2 was determined and suggests the use of these genes as biomarkers of the presence of CAC in patients with UC. Kuester et al. [90] demonstrated the hypermethylation of death-associated protein kinase (DAPK), a proapoptotic protein implicated in various apoptotic cascades, in UC-associated carcinogenesis in patients with long-standing disease. They also observed the overexpression of DAPK in inflamed colonic epithelium [90], indicative of a protective role for this protein. Thus, the inactivation of DAPK, mediated by promoter hypermethylation, might contribute to the accumulation of UC epithelial cells with genomic damage in response to inflammation and thus to the initiation of carcinogenesis and CAC.
The increasing evidence of inflammation-associated carcinogenesis includes a role for the key mediators of inflammation-induced DNA methylation: oxidative stress and increased levels of the pro-inflammatory cytokines, including IL-6, IL-β, TNF-α, and interferon-ɤ [17, 91–93]. The mechanisms by which these pro-inflammatory mediators alter DNA methylation during inflammation are not completely understood, but recent research has provided several insights. First, IL-6, the expression of which is increased during CAC, stabilizes DNMT1 protein levels in human colon cancer epithelial cells [17]. Second, interferon-ɤ was shown to increase nuclear 5’methylcytidine in human intestinal epithelial cell cultures by increasing DNMT3B mRNA levels [91]. Third, in an animal model of colitis, IL-1β expression was concordant with methylation induction, a phenomenon that is also observed in Helicobacter pylori infection, in which increases in IL-1β and TNF-α levels occur in parallel with the temporal changes in methylation levels [93, 94]. Finally, in a colon cancer cell line exposed to oxidative stress, DNMT1 and repressive factors were recruited to GC-rich regions of the genome [92]. These findings suggest that factors produced during inflammation alter mediators of DNA methylation. Thus, the aberrant DNA methylation patterns, rather than being a response to chromosomal insult, may in themselves promote an inflammatory environment. This is supported by a mouse model of colitis, in which aberrant DNA methylation occurred in the absence of macroscopic tumors and gradually increased until tumors developed [94].
According to this scenario, the duration of inflammation is an important factor in aberrant DNA methylation, which is consistent with the duration of IBD being a risk factor for the development of CAC. Whether the inflammation-induced increase in DNA methylation is targeted to specific regions of the genome or affects the genome as a whole remains to be determined.
5.4.4 MicroRNA Alterations
MicroRNAs (miRNAs) are 19–24 nucleotides long and serve as major regulators of gene expression, by targeting mRNAs post-transcriptionally [95]. Important roles for miRNAs have been confirmed in cellular differentiation, development, proliferation, and apoptosis. However, in cancer, these processes are deregulated, which implies that miRNAs are involved in carcinogenesis, perhaps at the tumor initiation and progression stages [96].
Indeed, increasing evidence suggests that miRNAs are involved in the carcinogenesis underlying CAC. Ludwig et al. [97] reported the upregulation of miR-21 in IBD-associated dysplastic lesions but not in tissues from patients with active IBD. The increase in miR-21 correlated inversely with the expression of PDCD4, a newly characterized tumor suppressor gene. Olaru et al. [98, 99] found that miR-224 and miR-31 expression increased successively at each stage of IBD progression, from non-inflamed to inflamed nonneoplastic, dysplastic, and finally cancerous mucosa. Both miR-224 and miR-31 levels could accurately discriminate normal or chronically inflamed IBD tissues from cancers. The authors also showed that miR-224 was involved in cell cycle regulation by targeting p21 and that miR-31, by targeting the negative repressor of hypoxia-inducible factor 1, regulated tumor angiogenesis, which would link both of these miRNAs to IBD-associated carcinogenesis.
In general, tumor-specific miRNA expression profiles are more informative and discriminatory than mRNA profiles. Furthermore, circulating miRNAs are highly resistant to RNase activity, unlike mRNA [100], which recommends their use as noninvasive biomarkers in the diagnosis of CAC. However, miRNA-based markers that confidently identify UC patients at increased risk of neoplasia have yet to be developed.
5.5 Molecular Markers to Identify the Risk of Neoplasia in UC Patients
CAC is a major cause of mortality in patients with UC [101, 102], such that diagnosis at an early or precancerous stage is crucial. A predisposition to colorectal neoplasia in UC is generally considered to depend on the diagnosis of UC at a young age and the presence of extensive colitis [103]. The prevalence of CAC in patients with UC is 8 % at 20 years after the initial UC diagnosis and increases to 18 % at 30 years [104]. Thus, surveillance colonoscopy with multiple random biopsies has been widely recommended for patients with long-standing and extensive UC [105]. However, because CAC is often difficult to detect endoscopically and its discrimination from inflammatory regenerative epithelium is histologically challenging, whether conventional surveillance colonoscopy is effective for the early detection of CAC remains a matter of contention. In addition, recent analysis demonstrated that the low yield and lack of clinical consequences from random biopsies in this high-risk population raise questions about the necessity and cost-effectiveness of routine random biopsy during UC surveillance [106]. Consequently, more accurate diagnostic modalities, such as chromoendoscopy and magnifying endoscopy, to identify potential sites of neoplasia in a nonneoplastic inflamed epithelium, together with analysis of p53 alterations, to distinguish neoplastic lesions from regenerative epithelium, have been evaluated [107, 108]. However, the labor-intensive nature and expense of these adjunctive modalities preclude their use in the surveillance of all UC patients with long-standing and extensive colitis. Rather, within this subgroup of patients, the ability to distinguish those who are at low vs. high risk of colorectal neoplasia would allow physicians to identify patients most likely to benefit from these more extensive screening methods.
5.5.1 Molecular Changes in the Nonneoplastic Mucosa in UC
The nonneoplastic mucosa of UC patients with CAC exhibits several molecular alterations (CIN, MSI, DNA aneuploidy, DNA methylation, telomere shortening, and gene expression), collectively referred to as a “field effect.” These alterations in the nonneoplastic UC mucosa may be promising biomarkers that allow the identification of UC patients at high risk of CAC.
5.5.2 Utility of Age-Related Methylation in Identifying CAC
In most human cancers, aberrant hypermethylation of promoter CpG islands, leading to the inactivation of key tumor suppressor genes, is a frequent and early step in tumorigenesis. In CRC, this epigenetic alteration is associated with two distinct subsets of genes: those that display a cancer-restricted methylation pattern (type C) and those that become methylated during aging (type A) [109]. Issa et al. [110] showed that the estrogen receptor (ER) of the normal colorectal mucosa becomes increasingly methylated with age and that hypermethylation of the ER gene is seen in most cases of sporadic CRC. Thus, age-related methylation may be an important contributor to the acquired predisposition to colorectal neoplasia and together with cancer-restricted methylation may be a feature of chronic inflammation-associated disease, including UC.
Issa et al. [86] were the first to demonstrate that the ER, MYOD, and CSPG2 genes and exon 1 of the p16 gene, all of which undergo age-related methylation in the colorectal mucosa, are intensively methylated in the high-grade dysplasia/CAC tissues of UC patients. Furthermore, they showed that these genes were also highly methylated in the nonneoplastic mucosa of these same UC patients. Accordingly, they proposed the use of age-related methylation as a molecular marker to identify UC patients at increased risk of developing dysplasia/CAC. This approach received support from another group that elegantly demonstrated high levels of ER gene methylation not only in regions of dysplasia/CAC but also in other regions widely scattered throughout the colorectum. The implication of the latter result is that analysis of a single biopsy sample (e.g., rectal biopsy) may suffice to identify higher-risk patients and that a large number of biopsy samples and total colonoscopy may not always be necessary [111, 112].
5.5.3 Methylation of the Putative Promoter Regions of miRNAs as a Marker of CAC in UC Patients
In an attempt to clarify whether age-related DNA methylation in nonneoplastic epithelium is an indicator of an increased risk for CAC, we selected miR-124, miR-137, and miR-34b/c as candidate genes, since their methylation is associated with increasing age and a field effect has been reported in the uninvolved colonic mucosa of patients with sporadic CRC [113–117]. We hypothesized that aberrant hypermethylation of the genes encoding these specific miRNAs in the normal, aging colorectal epithelium is an early event in CAC. Our systematic evaluation demonstrated the feasibility of using the methylation status of the miR-124, miR-137, and miR-34b/c genes as a promising biomarker in UC-associated neoplasia. When these biomarkers are used alone or in conjunction with the current guidelines for the diagnosis of UC-associated neoplasia, many of the current clinical challenges for managing these patients could be overcome. More importantly, the analysis of these biomarkers from a single rectal biopsy specimen has robust predictive potential in identifying UC patients at high risk for neoplasia elsewhere in the colorectum [118].
5.6 Conclusions and Perspectives
This chapter provided a review of the relationship between colonic inflammation and various tumor genetic events leading to CAC. The sequence of events ending in tumor formation is quite different from the events that give rise to the development of sporadic CRC. The early events in CAC involve DNA methylation, which regulates the expression of onco-suppressor genes, as well as mutation of p53, aneuploidy, and MSI. Tumor- and age-dependent methylation also occur in the nonneoplastic mucosa of UC patients with CAC, so-called field effects. A better understanding of the mechanisms for inflammation-induced carcinogenesis could identify IBD patients at high risk for CAC.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree