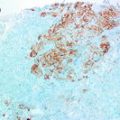
Fig. 3.1
H&E section from a mixed culture of fibroblast, breast epithelial cells and myoepithelial cells grown in a 3D collagen 1 matrix. Note particularly the organisation of clear myoepithelial cells around lumenal cells with the recapitulation of the histology of an acinus, towards the mid/bottom of this photomicrograph. The group of epithelial cells superior to it are not encompassed by myoepithelial cells and lack this organisation
As discussed, the most commonly used representative of normal luminal breast epithelium is MCF10A cells. Culture of these cells in MatrigelTM has yielded some impressive acini-like structures which are polarised, contain hollow lumens and produce basement membrane proteins [40, 59]. This has also been recapitulated in collagen I matrix [60]. Branching 3D duct-like structures have also been achieved through culture of HB2 cells in collagen I [51]. However, while culture of these cells in 3D matrix has resulted in structures akin to in vivo breast acini and ducts which cannot be achieved through culture in 2D, in vivo breast architecture is much more complex including a variety of other cell types that influence epithelial architecture. This has led to a rise in 3D co-culture with stromal cells with co-culture with fibroblasts most commonly reported [38, 47, 60] and examples of combinations of fibroblasts and adipocytes also demonstrated [61, 62]. It has emerged that fibroblasts regulate epithelial cell invasion [63] secreting various growth factors such as MMPs [64], which have been proven through use of 3D co-cultures. Markedly, some of these effects are only evident upon culture in collagen I [65]. Myoepithelial cells also play a key role in normal in vivo breast. Given their tumour suppressive nature [66, 67] and their capacity to maintain luminal cell polarity [68], it seems apt to include these cells in normal in vitro models of breast; however, to date studies incorporating these cells are lacking.
It stands to reason that luminal epithelial organisation is maintained by a delicate balance between the opposing functions of fibroblasts and myoepithelial cells. It seems apt that both these cell types are included in a 3D in vitro model of normal breast with luminal epithelial cells to accurately reflect the in vivo environment. One such tri-culture model has been achieved previously by Holliday et al. [38] but this used aberrant luminal epithelial cells thus representing DCIS. In our laboratory, we have developed a tri-culture model of normal breast that incorporates HB2 cells, myoepithelial cells and fibroblasts isolated from breast reduction mammoplasty samples in the physiologically relevant matrix collagen I. We have demonstrated that each individual cell type retained an in vivo phenotype and that combined shared morphology and phenotype with normal breast reduction mammoplasty acini. In addition, we have proved through overexpression of HER2 that formation of structures that accurately reflect DCIS in vivo can be achieved. What is more, we have proved that these structures are quantifiable by standard inexpensive laboratory techniques [unpublished].
However, despite better recapitulating the in vivo breast environment, models such as these are still reliant on cell line culture which may already represent aberrant breast epithelium. As discussed previously, the use of new primary cell cultures that represent the morphology, immunoprofile and genetic profile of normal and earlier benign breast lesions would be necessary to accurately model breast cancer initiation in the laboratory. The culture of primary breast epithelial cells in 3D MatrigelTM has been achieved previously [69] but has been limited to a single cell population. In order to accurately recapitulate the in vivo breast environment, multiple primary cell types would need to be isolated, characterised to ensure they accurately represented their in vivo counterparts, labelled to enable tracking and then cultured together in 3D ECM. The finite life span of primary cell cultures in vitro would make this challenging for researchers with further genetic and proteomic manipulation even more difficult.
Conclusion
In summary, when cultured under the right conditions, current available representative cell lines of normal and DCIS breast epithelium could provide valuable insight into the morphological, phenotypical and architectural changes that occur in the development from normal breast to benign lesions and cancer initiation. The use of cell lines grown in 2D offers the opportunity to study the complex intracellular mechanisms that may drive these processes in great detail while culture in 3D can perhaps better mimic the pathology and immunoprofile of these lesions in a more physiologically relevant setting. However, in order to recapitulate the genomic alterations that appear to drive the transition from normal breast to DCIS, better primary cell cultures would need to be developed.
Key Points
To understand the molecular pathology of breast cancer initiation and progression, appropriate models that allow experimentation in a controlled laboratory environment and reflect the heterogeneity of breast cancer are necessary.
Many cell lines used in breast cancer been virally immortalised and may therefore not be a true representative of cells in vivo.
To the best of the authors’ knowledge, there are no cell lines available that represent low grade DCIS or early breast lesions.
A limitation of all 3D in vitro models is the lack of a complex in vivo system complete with blood supply, immune infiltrates and regulation by hormonal cues.
In order to more faithfully recapitulate breast-like structures in a 3D matrix co-cultures with other cell types, notably fibroblasts and myofibroblasts, is necessary.
References
1.
Curtis C, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52.PubMedCentralPubMed
2.
Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Investig. 2011;121(7):2750–67.PubMedCentralCrossRefPubMed
3.
Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U.S.A. 2001;98(19):10869–74.PubMedCentralCrossRefPubMed
4.
Eccles SA, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15:5.
5.
Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55(2):231–73.PubMed
6.
7.
8.
9.
10.
Moinfar F, et al. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res. 2000;60(9):2562–6.PubMed
11.
12.
13.
14.
Forozan F, et al. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60(16):4519–25.PubMed
15.
Weigelt B, Bissell M. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol. 2008;18(5):311–21.PubMedCentralCrossRefPubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree