Laboratory Characteristics and Antibiotic Resistance of MRSA
S. aureus is a gram-positive, nonmotile, facultatively anaerobic coccus. In the laboratory, staphylococci tend to grow in grapelike clusters of cells, hence the genus name Staphylococcus, which is derived from the Greek word staphylé or “bunch of grapes.” The species name, aureus, Latin for golden, describes the color of S. aureus colonies growing in culture. Unlike many other bacteria, staphylococci can grow in environments with high concentrations of salt. This feature is commonly used to assist in the laboratory identification of Staphylococcus species. The presence of catalase activity can be used to distinguish staphylococcal species from several other genera of gram-positive cocci, including Streptococcus and Enterococcus. The production of the enzyme coagulase differentiates S. aureus from the other staphylococcal species (ie, the coagulasenegative staphylococci). Similarly, mannitol fermentation can also differentiate S. aureus from most other staphylococcal species.
The antibacterial effect of β-lactam antibiotics is the result of inhibition of penicillin-binding proteins (PBPs), which are bacterial proteins acting as catalysts of cell wall assembly. In MRSA, a single genetic change—acquisition of a gene encoding a variant penicillin-binding protein—confers resistance to nearly all β-lactam antibiotics. In this section, we discuss antibiotic resistance and laboratory characteristics of MRSA. The laboratory detection of MRSA and strain typing methods, both of which are important
components of MRSA surveillance programs and outbreak investigations, are also discussed here briefly.
Laboratory Definition of MRSA and Genetics of Resistance to β-Lactam Antibiotics The antibacterial effect of β-lactam antibiotics is thought to be mediated primarily through the inhibition of penicillin-binding proteins (PBP), which are bacterial proteins that act as catalysts of cell wall assembly. The main inhibitory target for β-lactam antibiotics is PBP2. While simple penicillin resistance in
S. aureus is due to a plasmid-encoded penicillinase that cleaves penicillin alone, resistance to methicillin is a result of acquisition by
S. aureus of a homologue of PBP2, designated as PBP2a, which has very low affinity for most β-lactams. Although typically referred to as MRSA, these strains are resistant not only to the antistaphylococcal penicillins (such as methicillin, nafcillin, and oxacillin) but also to all other currently available β-lactam antibiotics (with the exception of ceftaroline), including the first- through fourth-generation cephalosporins and the carbapenems.
3,4
PBP2a is encoded by the
mecA gene, which is located together with other functional genes within mobile staphylococcal cassette chromosomal (SCC
mec) elements. The SCC
mec has been categorized into multiple types based on genetic diversity, but contains three basic elements: the
mec gene complex (including the
mec gene and its regulatory elements), the
ccr gene complex (recombinase genes), and the joining regions (J regions).
2 Divergent
mecA genes that also code for low-affinity PBP2, which are designated as
mecB and
mecC, have been identified in humans, albeit uncommonly. Studies to date indicate that strains that carry
mecB and
mecC are detected as MRSA by most phenotypic tests but may be missed by molecular assays such as PCR that are specific for the
mecA gene.
5 The
mecB-mediated resistance is particularly concerning, as this gene has been identified on a mobile plasmid that harbors additional genes that encode resistance to aminoglycosides and macrolides.
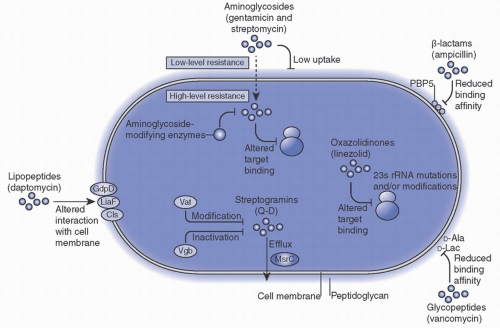
6Resistance to Non-β-Lactam Antibiotics Most strains of healthcare-associated (HA)-MRSA are also resistant to one or more other classes of antimicrobial agents, and multidrug resistance is common. This can be the result of mutations in chromosomal DNA or to acquisition of exogenous antibiotic resistance genes. In many instances, these additional resistance determinants may reside within SCC
-mec. Common mechanisms of resistance to non-β-lactam antibiotics are discussed briefly here and summarized in
Table 19-1.
Aminoglycoside resistance nullifies synergism between an aminoglycoside and cell wall active agents. It is mediated by acquisition of cytoplasmic aminoglycoside modifying enzymes (AME) that prevent the drug from binding to the ribosome. These genes are carried by mobile genetic elements including transposons (
aacA-aphD, aphA) or by plasmids integrated into the SCCmecII (
aadD).
3,4Fluoroquinolones target the DNA gyrase and topoisomerase IV in
S. aureus. Resistance has been demonstrated through mutations to topoisomerase IV (
grIA) or DNA gyrase (
gyrA) that reduce drug binding and expression of efflux pumps (
NorA, NorB, NorC). Topoisomerase mutants often develop in the setting of subinhibitory concentrations of quinolones.
3,4Tetracycline resistance is typically mediated through efflux pumps or ribosomal protection.
Tet efflux pump genes,
TetA(K) and
TetA(L), are part of the SCC
mecIII cassette.
TetO and
TetM are located on a conjugative transposon and encode a protein with GTPase activity that confers ribosomal protection. Resistance to the related glycylcycline antibiotic, tigecycline, has also been reported rarely.
3,4Trimethoprim, typically used in combination with sulfamethoxazole (TMP-SMX), is a folic acid metabolism inhibitor of the enzyme dihydrofolate reductase (DHFR). This enzyme is essential for bacterial survival. Intermediate resistance has been described by chromosomal mutation in DHFR (
dfrB), but high-level resistance is due to horizontal acquisition of genes coding for mutant enzymes (
DfrA, DfrK, DfrG).
3,4Daptomycin is a cyclic peptide antibiotic that is used widely to treat serious MRSA infections. It functions by disrupting the bacterial cell wall, resulting in depolarization and cell death. Resistance to daptomycin can occur following prolonged courses of daptomycin, and is accelerated by prior treatment with vancomycin. Mutations associated with vancomycin-intermediate
S. aureus (VISA) can also reduce susceptibility to daptomycin. The most common mutation to confer daptomycin resistance results in increased charge of the outer face of the membrane (
mrpF), although several other mechanisms have been described.
3,4Linezolid is a synthetic oxazolidinone that is used in difficult-to-treat MRSA infections. A second-generation drug in this class, tedizolid, has also been approved for the treatment of skin and soft tissue infections. Resistance to these drugs has been described rarely. Resistance mechanisms include acquisition of a methyltransferase (
Cfr, OptrA) resulting in reduced linezolid binding or alteration of the 23S ribosomal RNA at the linezolid binding site.
3,4Vancomycin and Other Glycopeptide Resistance Vancomycin is a glycopeptide antibiotic that is commonly used to treat infections caused by MRSA in hospitalized patients. Semisynthetic lipoglycopeptides related to vancomycin have also been approved for treatment of MRSA, including oritavancin and telavancin. Vancomycin-resistant
S. aureus has been described rarely, usually after prolonged vancomycin treatment.
3,4
In
S. aureus, intermediate resistance to the glycopeptides is due to mutations in the bacterial chromosome. These mutations cause changes in the structure of the peptidoglycan component of the cell wall, leading to a thicker wall with more un-crosslinked D-alanyl-D-alanine (D-ala-D-ala) terminals. These excess D-ala-D-ala terminals bind to glycopeptide molecules and prevent them from reaching their intended target. Although the terms VISA and glycopeptide-intermediate
S. aureus (GISA) are often used interchangeably, some VISA isolates retain
in vitro susceptibility to the glycopeptide teicoplanin. Heteroresistant VISA populations (h-VISA) have also been described.
3,4One of the most feared scenarios has been the development of high-level vancomycin resistance in
S. aureus (VRSA) due to acquisition of the plasmid-mediated vancomycin resistance gene,
vanA, from vancomycin-resistant
Enterococcus (VRE). The mechanism of high-level vancomycin resistance involves alteration of the peptidoglycan synthesis pathway; it is discussed in more detail later in this chapter. The first clinical isolate of VRSA was identified in the United States in 2002. Since then, 14 additional cases have been described.
7 Commonalities identified among most of the reported cases include prior history of VRE colonization or infection, prior history of MRSA colonization or infection, and prior receipt of vancomycin therapy.
8 The failure of VRSA strains to expand and disseminate remains poorly understood, but there is some evidence that carriage of
vanA confers a fitness cost. It has been shown that in the absence of vancomycin, VRSA growth rates are similar to MRSA strains. However, induction of
vanA by exposure to vancomycin results in significant growth reduction of VRSA.
9MRSA Virulence Factors MRSA has the potential to carry a variety of virulence factors. Many of these factors are acquired by horizontal transfer of mobile genetic elements. A number of toxins have been described including hemolysins, leukocidins, enterotoxins, toxic shock syndrome toxin 1 (TSST-1), and exfoliative toxins. Enzymes to promote tissue invasion (eg, hyaluronidase) and factors to evade immune response (eg, protein A) also contribute to the pathogenicity of MRSA. Some of the most studied factors include arginine catabolic mobile element (ACME) and Panton-Valentine leukocidin (PVL); genes that encode these factors are carried on mobile genetic elements found mostly in USA300 MRSA strains. Additionally, genomic islands can carry a wide variety of mobile virulence factors that remain stable following horizontal gene transfer.
1 An in-depth discussion of virulence factors is outside of the scope of this chapter but can be found in recent reviews.
1,2
Laboratory Methods for Detection and Identification of MRSA Contemporary clinical microbiology laboratories can choose from among a wide range of accurate, simple, commercially available detection and identification methods for MRSA. Culture-based direct detection methods include mannitol salt agar plus oxacillin and with or without other components, and various proprietary selective and differential chromogenic agars. Overnight broth enrichment has often been demonstrated to improve the yield of surveillance cultures. Methods to identify MRSA from isolated colonies include the oxacillin screen agar test, cefoxitin disk test, and the rapid PBP2a antigen test.
10 Commercial tests for the rapid molecular detection of genes specific for MRSA are also available; these tests can be used directly on clinical specimens, such as anterior nares swabs; on broth cultures; and on isolated colonies. Characteristics of some common tests are described below and summarized in
Table 19-2.
Phenotypic detection of MRSA Chromogenic agars allow for simple and relatively rapid identification of methicillinresistant strains of
S. aureus. Clinical or surveillance samples are plated onto selective agars that inhibit the growth of methicillin-susceptible strains of
S. aureus and produce specific color changes in colonies of MRSA. Results can be obtained in as little as 24 hours. Multiple formulations of chromogenic agar are available commercially with some differences in performance reported.
11,12
The cefoxitin disk diffusion screening test is a highly sensitive and specific phenotypic means of detecting
mecA-mediated methicillin resistance in
S. aureus culture. This test has mostly replaced the oxacillin screen agar test because cefoxitin is a more potent inducer of the
mecA
gene and resistance is less likely to be expressed heterogeneously than with oxacillin.
11,12 Both the cefoxitin disk test and the oxacillin screen agar test require overnight incubation of pure cultures of
S. aureus.
Latex agglutination and immunochromatographic assays are available that directly detect PBP2a in pure cultures of MRSA. These assays allow for rapid detection of resistance (<1 hour) and are considered the “gold standard” of phenotypic methods for identification of MRSA.
11,12Molecular detection of MRSA Nucleic acid amplification tests (NAAT) such as PCR are available for the detection of MRSA are available for screening of both clinical and surveillance specimens, including nasal and skin swabs. PCR-based assays are also approved for detection of MRSA from colony or broth culture specimens. As compared with culture-based methods, PCR is highly sensitive and specific for detection of
mecA-mediated methicillin resistance and is usually considered to be the “gold standard” molecular test for MRSA detection. Performance of these assays is dependent in part on the specific genetic sequence that is targeted within the SCC
mec cassette. Many assays have targeted the SCC
mec–
orfX junction that may also be present in methicillin-resistant coagulase-negative staphylococci, leading to false-positive results. Newer assays detect a combination of multiple genetic sequences to improve performance. As noted above,
mecB and
mecC may not be detected by PCR methods that are specific for
mecA. Advantages to PCR are primarily speed: definitive results are available within <1-3 hours of receipt of the specimen in the laboratory. The cost of commercial PCR tests for MRSA detection are typically higher than the cost of culture-based testing methods.
11,12
Rapid identification of MRSA with the use of PCR-based assays has implications for antimicrobial stewardship and may allow more rapid implementation of infection control measures designed to reduce the risk of MRSA transmission.
6,13 However, in order to achieve these outcomes, preanalytic and postanalytic factors must be optimized, such as rapid delivery of specimens to the laboratory for testing, and rapid release of test results to a healthcare provider who can change antibiotic orders or initiate infection control measures.
14 Utility of rapid MRSA identification to reduce transmission remains controversial and will be discussed later in this chapter.
11,12MRSA Strain Typing Strain characterization and typing of MRSA isolates is important to help delineate the epidemiology of transmission, and detection of virulence and antibiotic resistance markers can have clinical implications. Several historical strain typing methods and MRSA designations are listed in
Tables 19-3 and
19-4. Whole genome sequencing (WGS) has largely replaced these older typing methods, because of its greater discriminatory power and ability to identify chains of transmission as well as infection clusters. Analysis by WGS was cost prohibitive in the past, but now benchtop instruments and lower cost systems have made it more accessible to many laboratories. The biggest challenge for implementation of WGS remains the postsequencing workflow, which requires computing and bioinformatics knowledge that may not be available readily in clinical laboratories. WGS
is predicted to become increasingly more widely used, and has already proved to be a powerful tool in MRSA transmission and outbreak investigation.
2,15
Epidemiology
Prevalence of MRSA in Healthcare Settings MRSA is endemic in the majority of healthcare centers worldwide. In the United States, ˜50% of
S. aureus HAIs are caused by MRSA, with MRSA reported as an important cause of ventilator-associated pneumonia (VAP), surgical site infections (SSI), and central line-associated bloodstream infections (CLABSI).
16 A similar distribution of MRSA infection types is reported for intensive care unit (ICU)-acquired HAIs in Europe, with MRSA representing ˜30% of
S. aureus isolates, although there is substantial variability in prevalence between European countries.
17
In a retrospective review of
S. aureus among all hospital isolates reported to the U.S. National Nosocomial Infections Surveillance (NNIS) System, the percentage of MRSA increased from 2.4% in 1975 to 29% in 1991.
18 MRSA infections were initially reported to occur among those who frequented healthcare facilities (eg, hemodialysis units) or among those admitted into acute or long-term care facilities. However, MRSA became an increasingly common cause of community-onset and invasive staphylococcal infections during the next two decades, mostly attributed to clonal expansion of pulsed-field gel electrophoresis-defined USA300. MRSA is now a leading cause of both HAIs and community skin and soft tissue infections (SSTI).
16,19Recent reports reveal a decreasing overall prevalence of MRSA infections since the early to mid-2000s. This trend is demonstrated among multiple large geographic
cohorts in the United States, in Europe, and among both adult and pediatric populations.
20,21 A decrease in the overall number of hospitalizations related to all
S. aureus infections (including both MSSA and MRSA) was appreciated from 2010 to 2014, with a decrease of ˜15% in MRSA infections.
22 Total hospital-associated community-onset and hospital-onset MRSA infections decreased by ˜25%-50% from 2005 to 2011, with decreased prevalence of invasive MRSA infections, including CLABSI, also reported during this period.
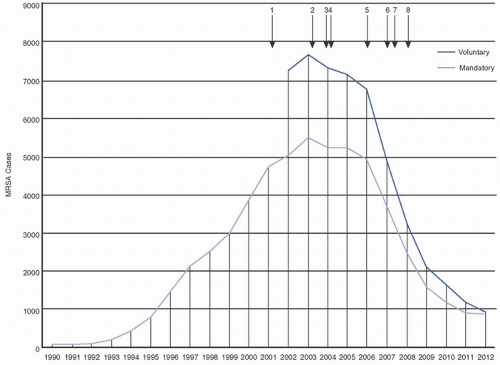
20,23,24,25 Duerden et al. (2011) demonstrated an 80% reduction in MRSA bloodstream infections (BSI) in England following implementation of mandatory reporting and targeted infection control and prevention initiatives (
Fig. 19-1).
21 The extent to which observed reductions in MRSA are due to targeted MRSA control efforts, global infection prevention efforts, and/or natural changes in MRSA prevalence over time is unclear.
26 Although the observed reductions in MRSA HAI are encouraging, it is important to realize that MRSA remains prevalent in most healthcare facilities.
Evolving Epidemiology of Community-Associated and Hospital-Associated MRSA Over the past two decades, the epidemiology of MRSA has changed as a result of clonal dissemination of novel MRSA strains that are genetically and epidemiologically distinct from typical HA-MRSA strains (
Table 19-5). Community-associated
MRSA (CA-MRSA) appears to have arisen as the result of migration of SCC
mec type IV and V into MSSA, with USA300 (ST8) as the dominant strain in the United States. CA-MRSA strains are often resistant only to β-lactam antibiotics and perhaps one or two additional antibiotic classes.
2 However, resistance to additional classes of antibiotics is being reported with increasing frequency.
Historically, the major determinant in characterizing an MRSA infection as either “healthcare associated” or “community associated” was the time of onset, or the time to identification of the infection after admission to the hospital. A recent epidemiologic classification scheme that acknowledges the ongoing risk of healthcare exposure even after discharge from a healthcare facility is shown in
Table 19-6. As noted earlier, though, molecular strain typing has also been used to attempt to distinguish healthcare- and community-associated risks of MRSA infections. The ability to reliably distinguish the likely sources of MRSA strains has become more difficult as strains of MRSA that were traditionally considered to be CA-MRSA, such as USA300, have become established in hospitals and HA-MRSA strains have entered the community.
27Nosocomial outbreaks of CA-MRSA have been reported since 2003.
2 In some institutions, USA300 is now the major nosocomial clone of invasive disease, replacing USA100 as the predominant isolate.
28 Complex local community and hospital transmission networks of USA300 have been identified, with the community as the primary driver of MRSA BSI.
29 From 2005 to 2013, the incidence of BSI caused by USA100 declined by >60%, with >80% decline in hospital-onset USA100 BSI. USA100 continues to be associated with classical healthcare exposures (ie, central venous catheters, prior hospitalization) despite the expansion of USA300.
30 A full discussion of the evolving epidemiology of MRSA clones is beyond the scope of this chapter, but excellent reviews can be found in the literature.
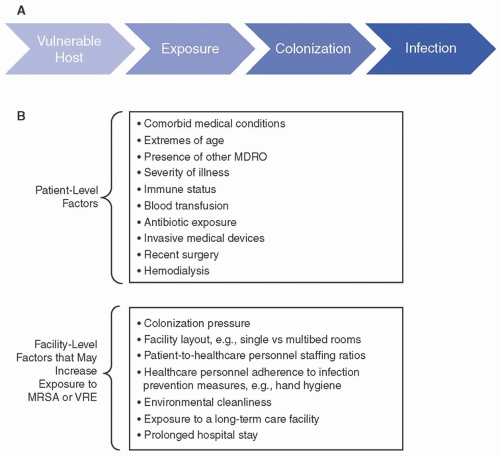
2Risk factors for MRSA MRSA infection is the result of a complex interplay of multiple factors, including those specific to the host (eg, comorbid conditions, sex, age, immune defenses, exposure to antibiotics or invasive medical devices), the institution (eg, colonization pressure, adherence of healthcare providers with hand hygiene and other prevention measures). Many of the patient-level and facility-level risk factors for development of nosocomial MRSA infection are common to other multidrug-resistant organisms (MDROs). As is generally true for other MDROs, MRSA colonization typically precedes infection (
Fig. 19-2). MRSA colonization itself is a major risk factor for MRSA infection. Here, we discuss risk factors for MRSA at each step from acquisition of colonization to development of MRSA infection.
Patient-level risk factors for MRSA A number of host factors have been identified that increase the risk of MRSA acquisition, establishment of colonization, and progression to MRSA infection (
Fig. 19-2). Due to overlap, it is difficult to discern which factors contribute the most to each step in the pathway toward MRSA infection. In addition, studies of MRSA infection may not have surveilled for preceding colonization, thus making it difficult to distinguish risk factors specific for colonization versus infection.
Commonly reported risk factors for MRSA colonization in the host include certain underlying comorbidities such as diabetes, extremes of age, open skin lesions, chronic renal insufficiency, neurologic disorders (prior stroke, dementia, hemiplegia), human immunodeficiency virus (HIV) infection, and cystic fibrosis. Receipt of antibiotics, the presence of an invasive medical device (such as indwelling urinary, vascular catheters, tracheostomy tubes, and feeding tubes) recent surgery, or colonization with another MDRO are also associated with increased risk of MRSA. Facility-associated risk factors for MRSA colonization include high MRSA
colonization pressure, admission from a long-term care facility, and prolonged hospitalization.
1,31,32,33,34,35,36 These and other risk factors are shown in
Figure 19-2.
MRSA Reservoirs in Healthcare Facilities A variety of factors within the healthcare system and the healthcare delivery process have been implicated in the acquisition or transmission of MRSA. The most important MRSA reservoir in the acute care setting is patients who are colonized or infected with MRSA. The risk of acquisition of MRSA during hospitalization increases as the prevalence of MRSA among hospital patients increases (designated as “colonization pressure.”).
37 In one study, it was observed that when the weekly colonization pressure exceeded 30%, the risk of MRSA acquisition was five times higher than when colonization pressure was <10%.
38 It should be noted, however, that because MRSA can asymptomatically colonize patients, the true prevalence of MRSA in most healthcare facilities is often not known unless MRSA screening is performed. MRSA prevalence or colonization pressure in healthcare facilities varies substantially. In one study, a range of 3.7%-20% was observed in MRSA prevalence across multiple ICUs.
33
Contamination of environmental surfaces and healthcare equipment in the rooms of MRSA colonized or infected patients is relatively common. Once in the environment,
S. aureus can persist for extended periods.
39 In one study, an average environmental contamination of >20% was found on numerous high-touch hospital surfaces.
40 HA-MRSA strains were more often identified on surfaces in contact with hospital healthcare personnel (HCP) (ie, medicine room, chart holders, access doors, medicine cart), while public surfaces (ie, hand rails, coffee machine, elevator) had equal detection of both HA- and CA-MRSA strains (ie, USA100 and USA300).
40 Numerous environmental foci have been implicated in MRSA contamination, including the air, clinical tools (ie, stethoscopes, blood pressure cuffs), bedding equipment (ie, mattress, bed wheels), furniture, sinks, toilets, HCP and patient clothing, and stationary utensils (ie, pens, keyboards).
39 Thus, thorough environmental cleaning of both high-touch and common area surfaces is considered an important component in the control of the spread of MRSA in the healthcare setting.
HCP may transmit MRSA indirectly from patient to patient, or indirectly from an environmental source to a patient, through transient contamination of their hands or clothing. Less commonly, HCP may transmit MRSA directly from a colonized site on their own body to a patient. Rarely, airborne transmission of
S. aureus from nasally colonized HCP, so-called cloud disseminators, has also been epidemiologically linked to hospital outbreaks. The risk of
S. aureus airborne transmission has been associated with viral upper respiratory infection.
41 In a meta-analysis of
127 published studies, the average MRSA prevalence worldwide among HCP was found to be 4.6%.
42 A separate meta-analysis of 31 studies from the United States and Europe found a lower overall prevalence of 1.8%, although when one study from the Netherlands with very low rates of colonization was excluded the rate increased to 4.4%.
43 Risk factors for MRSA carriage by HCP include cutaneous lesions or skin conditions, sinusitis/rhinitis, recent antibiotic use, employment in areas with high patient MRSA prevalence, close contact with patients, and poor attention to infection control. In one study, the risk of colonization in nurses was nearly twofold higher than in other healthcare staff; the study’s authors hypothesized that this finding was explained by nurses’ more frequent and close contact with patients.
43 Routine MRSA screening and decolonization of HCP is not recommended currently. However, treatment of colonized HCP implicated in MRSA outbreaks has been an important step in outbreak control.
44MRSA colonization in acute care hospital patients S. aureus is a common component of the indigenous microbiota of humans and many animals. Population-based studies suggest that approximately one-third of the population is asymptomatically colonized with
S. aureus, with about 1.3%-1.5% colonized with MRSA.
45,46 The overall prevalence of U.S.
S. aureus nasal colonization decreased from 2001 to 2004, but the proportion with MRSA increased during that interval from 0.8% to 1.5%. Persistent or transient carriage of MRSA is most commonly detected in the anterior nares, but carriage on other mucous membranes and skin is also detected. Common sites of cutaneous carriage include the axilla, groin, perianal and perineal areas, wounds and sites of chronic skin disease, as well as foreign body exit sites.
47,48 Inguinal or perirectal colonization is more common in men than in women, with men who have sex with men disproportionately represented.
48 If screening is not performed, it is estimated that >50% of asymptomatically colonized patients would be missed.
33 Approximately 20% of patients remain persistently colonized with MRSA, while 60% are only intermittently colonized. Both intermittent and persistent colonization increase subsequent risk of invasive MRSA infection.
47
The duration of MRSA colonization is not well defined, and may be prolonged in certain patient populations. Persistent carriage is associated with ongoing residence in a healthcare institution, immunosuppressive therapy, hemodialysis status, skin disease, and presence of decubitus ulcers. In one prospective study, nearly 50% of hospitalized patients remained colonized at 1 year and ˜20% at 4 years.
49 The estimated half-life of MRSA colonization was 40 months in one earlier study,
50 with 50% clearance appreciated at 88 weeks in a separate recent analysis.
51MRSA colonization and reservoirs in long-term care facilities MRSA has emerged as an important microorganism in postacute long-term care facilities (LTCF). Epidemiologic descriptions of MRSA in these facilities are heterogeneous, likely reflecting the heterogeneity of the patient populations. Residents of these facilities are important reservoirs for MRSA transmission within the healthcare system.
52 Transmission of MRSA within the healthcare network is complex, with bidirectional movement of MRSA between acute and long-term care facilities.
52,53
A wide range of MRSA prevalence rates in long-term care facilities have been reported. In one study of MRSA nares colonization in elderly patients admitted to LTCF in the United States, the overall prevalence was 20%, but there was significant variation between facilities.
54 Some cohorts have even described prevalence rates exceeding 50%.
55Risk factors for MRSA carriage in LTCF include antibiotic exposure, bed-bound status, recent hospitalization, severe comorbidities, presence of wound or chronic skin disease, and presence of medical devices.
56 Decreased risk for MRSA carriage has been associated with MRSA decolonization following hospital discharge, use of infection prevention interventions within the facility, and increased HCP-to-patient ratio.
57,58MRSA colonization and reservoirs in the community CA-MRSA now serves as an important reservoir for importation of MRSA into the hospital. Skin and soft tissue infections are the most common type of infection caused by CA-MRSA, but other manifestations include pneumonia, BSI, ear infections, and joint infections.
59 Risk factors for CA-MRSA infection include extremes of age (<2 years or >65 years), HIV infection, athletic activity, injection drug use, male sexual activity, active military service, incarceration, residence in a homeless shelter, and close contact with a person colonized or infected with MRSA.
60,61
MRSA Transmission In healthcare settings, MRSA is generally spread through contact with contaminated HCP hands, fomites, or medical equipment. Use of WGS within an English ICU revealed that ˜20% of MRSA acquisitions could be explained by transmission between patients who were coresident in the ICU. The authors also demonstrated related isolates in patients who were not admitted to the ICU concurrently, suggesting that some acquisitions may have occurred outside the ICU or from other nosocomial sources.
62 In a separate large integrated epidemiologic analysis, Coll et al.
63 found that more than half of cases of hospital MRSA transmission with epidemiologic and bacterial genomic links were part of transmission clusters with hospital contacts, most frequently on the same ward. Patterns of transmissions within certain hospital wards, as well as movement of specific MRSA-infected individuals through multiple hospital units, were both observed.
63
Transmission dynamics of MRSA in the community are complex and appear to have to have important geographic connotations.
63 Infection prevention of MRSA within the community is an area of ongoing research and has important implications for the spread of MRSA isolates within both the community and hospital setting.
Clinical Manifestations of Healthcare-Associated Infections Caused by MRSA
Bloodstream Infection (Including CLABSI) and Infective Endocarditis S. aureus is a leading cause of BSI worldwide and was the second most common cause of CLABSI in the United States from 2011 to 2014. Of these isolates, >50% were MRSA.
16,77 The prevalence of MRSA bacteremia is highly variable and dependent on geographic and individual patient population factors, with estimates ranging from 1% to >50%.
78 For patients with hospital-onset MRSA infection, bloodstream infection was the most common presentation.
20
Complications of
S. aureus BSI are common, estimated at 11%-53%. Predictors of complications include persistent positive blood cultures at 48-96 hours, community acquisition, skin findings suggesting acute systemic infection, and persistent fever. Patients with immunocompromise, such as HIV infection or malignancy, are at higher risk of complications. Nearly any body site can be seeded during bacteremia, with infective endocarditis as a common complication. Patients with prosthetic valves are at higher risk of MRSA infective endocarditis compared to patients with native valves.
79Healthcare-Associated Pneumonia MRSA is an important cause of nosocomial pneumonia, including hospital-acquired pneumonia (HAP) and ventilator-associated pneumonia (VAP).
16 Current guidelines recommend utilizing hospital antibiogram data to reduce the unnecessary use of empiric anti-MRSA antibiotic treatment for nosocomial pneumonia. An active agent against MRSA for the empiric treatment of suspected nosocomial pneumonia is only recommended in patients with risk factors for antimicrobial resistance, high risk of mortality, or in patients in units with high prevalence (>10%-20%) of MRSA.
80 An important clinical predictor of MRSA nosocomial pneumonia is receipt of intravenous antibiotics within the previous 90 days. A positive MRSA screen from nasal or respiratory samples and residence in units where >20% of
S. aureus isolates are MRSA have also been suggested as a predictors of MRSA nosocomial pneumonia.
80 Diagnosis of MRSA nosocomial pneumonia can be difficult due to variability of symptoms and difficulty of obtaining a microbiologic diagnosis. However, negative nasal MRSA PCR screening has excellent negative predictive value for MRSA pneumonia and has been useful in guiding de-escalation of antibiotic therapy.
13
Skin and soft tissue infections (SSTI) and SSI S. aureus is the most common cause of SSTI and SSI and causes a wide variety of clinical manifestations, ranging from erysipelas to deep tissue infections and abscesses. Approximately 30% of breast, cardiac, neurological, prostate, and vascular HAI SSI and >40% of orthopedic HAI SSI were attributed to
S. aureus in the United States from 2011 to 2014. MRSA prevalence in these isolates was ˜40%.
16 Nasal and skin colonization are the major risk factors for development of complicated SSTI due to MRSA in hospitalized patients. Other risk factors include previous MRSA infection, recent antibiotic treatment, age >65 years, chronic illness (ie, diabetes, cardiovascular disease, chronic renal failure), HIV infection, or immunocompromised state.
81