Gene
Alteration
Frequency (%)
Histology
Smoking history frequent patterns
EGFR
Mutation
10–35
Adenocarcinoma, few adenosquamous carcinoma
Frequent in never smokers
KRAS
Mutation
15–25
Mostly in adenocarcinomas, frequently mucinous morphology
Former/current smokers, rarely never smokers
FGFR1
Amplification
20
More frequent in squamous cell carcinoma than adenocarcinoma
Former/current smokers
MET
Amplification
5–20
Acquired resistance to EGFR TKI—adenocarcinoma
Frequent in never smokers
MET
Amplification
2–4
Previously untreated—patients—adenocarcinoma
Frequent in never smokers
PTEN
Mutation
4–8
Squamous cell carcinoma
Ever smokers
DDR2
Mutation
~4
Adenocarcinoma and squamous cell carcinoma
No significant association yet described
ALK
Rearrangement
3–7
Adenocarcinoma, acinar histology, or signet-ring cells
Light smokers (<10 pack years) and/or never smokers
BRAF
Mutation
1–3
Mostly in adenocarcinomas
Former/current smokers
PIK3CA
Mutation
1–3
More frequent in squamous cell carcinoma than adenocarcinoma
Ever smokers and never smokers
NTRK1
Rearrangement
3
Adenocarcinoma
Never smokers
ERBB2 (HER2)
Mutation
2–4
Adenocarcinoma
Never smokers
ROS1
Rearrangement
2
Adenocarcinoma
Light smokers (<10 pack years) and/or never-smokers
AKT1 (AKT)
Mutation
1
Adenocarcinoma and squamous cell carcinoma
No significant association yet described
MAP2K1 (MEK1)
Mutation
1
Mostly in adenocarcinomas than squamous cell carcinoma
No significant association yet described
NRAS
Mutation
1
Adenocarcinoma
Former/current smokers
RET
Rearrangement
1
Adenocarcinoma
Frequent in never smokers
The first randomized trial involving East Asian patients with advanced lung adenocarcinoma (stage IIIB/IV), the Iressa Pan-Asia Study [IPASS], and subsequent five additional trials showed that for patients with an activating EGFR mutation, initial treatment with an EGFR TKI (gefitinib and/or erlotinib) was superior to standard platinum-based chemotherapy [12]. These findings have been a stepping stone for progress in selection of targeted therapy based on molecular genetic alterations in the tumors.
There are certain patterns noted in distribution of recurrent mutations. Never smokers with adenocarcinoma have the highest incidence of EGFR, HER2, ALK, RET, and ROS1 mutations while in smokers, adenocarcinomas with mucinous histology tend to have frequent KRAS mutations [6].
The clinical practice guidelines (CPG) were recently published for Lung Cancer Biomarkers to identify predictive biomarkers of molecular therapy for EGFR and ALK genes by collaborative efforts by College of American Pathologists (CAP), International Association for the Study of Lung Cancer (IASLC), and Association for Molecular Pathologists (AMP). Molecular testing for EGFR and ALK mutations is discussed below in detail. Other mutations that show promise for targeted therapies are currently in various stages of clinical trials and these will be discussed subsequently in this chapter. Next-generation sequencing (NGS) as testing platform is increasingly being used for detecting multiple mutations across many genes with a focused custom panel or a whole exome/whole genome approach. The NGS testing protocols and details are discussed elsewhere in this book. Each assay has their pros and cons and the summary of most widely used molecular testing methods is listed in Table 8.2.
Table 8.2
Types of frequently used molecular methods, mutations detected, and assay performance
Methods | Types of mutations | Assay performance | ||||||
|---|---|---|---|---|---|---|---|---|
Point mutation (SNVs) | Small insertions, deletions, duplications | Copy number changes (large deletion, insertions, amplification) | Structural alterations—translocations, inversions, etc. | Chemistry—detection | Sensitivity to detect % of mutant tumora | Specificitya | Turnaround timea | |
Sanger sequencing (traditional or enrichment: LNA, PNA) | XX | XX | XX (RNA) | Dideoxy PCR—CE | Low-average | High | 2–3 days | |
Allele-specific PCR | XX | Real-time PCR or CE | High | High | 1–2 days | |||
Pyrosequencing | XX | X | Pyrophosphate detection upon base incorporation | High | High | 1–2 days | ||
Single-base extension PCR | XX | X | Dideoxy PCR—CE | Average-high | High | 2–3 days | ||
Single-base extension PCR-MS | XX | X | MALDI-TOF | Average-high | High | 2–3 days | ||
MLPA | XX | XX | Ligation-PCR-CE | Average-low | High | 2–3 days | ||
In situ hybridization (FISH/CISH) | XX | XX | In situ hybridization—fluorescence microscopy | Low | Low | 2–3 days | ||
NGS—Targeted sequencing-amplicon capture | XX | XX | Dependent on platform used | Highb | Highb | Days to weeks | ||
NGS—Targeted sequencing-hybridization capture | XX | XX | XX | XX | Dependent on platform used | Highb | Highb | Days to weeks |
NGS—whole-exome sequencing | XX | XX | XX | X | Dependent on platform used | Highb | Highb | Weeks to months |
NGS—whole-genome sequencing | XX | XX | XX | XX | Dependent on platform used | Highb | Highb | Weeks to months |
Array CGH | XX | Competitive hybridization (FISH), scanningc | Average | Average | 1 week to 1 month | |||
Genome-wide SNP array | XX | XX | Competitive hybridization (FISH), scanningc | Low | Low | Less than 1 week | ||
Any applicable drugs currently approved or not approved and in different stages of clinical trials are listed at the end of this chapter in Appendix, in a table generated from Web site http://www.mycancergenome.org/content/disease/lung-cancer/.
Epidermal Growth Factor Receptor (EGFR)
EGFR (ERBB1) gene, located on chromosome 7p12, encodes for a protein which is one of the four receptor tyrosine kinases belonging to ERBB family receptor tyrosine kinases. Receptor binding with ligands such as epidermal growth factor (EGF), transforming growth factor, alpha (TGFA), amphiregulin (AREG), epithelial mitogen/epigen (EPGN), betacellulin (BTC), epiregulin (EREG), and heparin-binding EGF-like growth factor (HBEGF), triggers homo- and/or heterodimerization of the receptors leading to autophosphorylation on key cytoplasmic residues. This in turn activates complex downstream signaling cascades in at least four major downstream signaling pathways including the RAS-RAF-MEK-ERK, PI3K-AKT, PLCG1-PRRT2 (PLCgamma-PKC), and STATs modules and sometimes, G protein-coupled receptor signaling and NFKB1 (NF-kappa-B) signaling cascade. These activated signaling pathways drive many cellular responses such as changes in gene expression, cytoskeletal rearrangement, and anti-apoptosis and increased cell proliferation. Due to activating mutations in cancer cells, there is a constitutive or sustained active state that leads to pro-survival signaling which increases potential for invasiveness, proliferation, metastasis, and angiogenesis [14].
Mutations
In the USA about 10 % of patients with NSCLC tumors harbor EGFR mutations while the frequency is much higher in East Asia, about 35 % [15–17]. The clinicopathologic characteristics of EGFR mutations are as follows: they are more frequently found in tumors from females, and never smokers (defined as less than 100 cigarettes in a patient’s lifetime) with adenocarcinoma histology with particularly lepidic growth pattern. Other histological patterns of adenocarcinoma and some adenosquamous carcinomas may contain EGFR mutations. In addition, they are observed in other subsets of NSCLC, including in former and current smokers [15–17]. These mutations in lung adenocarcinoma are mutually exclusive with many other oncogenic mutations like KRAS, BRAF, and ALK gene rearrangement. The mutations occur within EGFR exons 18–21 encoding a portion of the EGFR kinase domain. EGFR mutations are usually heterozygous with frequent gene amplification of the mutant allele [18, 19]. Approximately 90 % of these mutations involve exon 19 (deletions) or exon 21 point mutation (L858R) [20]. These mutations increase the kinase activity of EGFR, leading to hyperactivation of downstream pro-survival signaling pathways [21].
Most of the EGFR mutations in exon 18–21 are predictive of sensitivity towards EGFR TKI such as gefitinib or erlotinib (see Table 8.2). EGFR exon 20 T790M mutation is associated with acquired and in rare cases primary resistance to EGFR TKI. Most exon 20 insertions have been shown to have reduced EGFR TKI response. However, the EGFR A763_Y764insFQEA mutation is an exception and has been associated in retrospective studies with increased EGFR TKI sensitivity [22]. Data remain very limited for some types of insertions. G719 and L861 mutations are considered somewhat less sensitive compared to major exon 19 and 21 mutations. Mutations at E709 and S768 often occur in combination with another of the listed mutations [23–29].
Role of the Molecular Tests
The EGFR mutation testing is not used as a diagnostic test for lung adenocarcinoma. Tumors with EGFR mutations in exons 18–21 lead to constitutive activation of the downstream signaling and this is thought to yield more aggressive tumor phenotypes. EGFR mutation testing is recommended for patients being considered TKI therapy. To that effect CPG for EGFR testing were published in 2013.
Briefly, after a systematic review of extensive published data on EGFR-mutated lung cancer and ALK rearrangements in lung cancers, CAP/IASLC/AMP with interest in the diagnosis and management of lung cancer published evidence-based recommendations for the molecular testing of lung cancers for these two predictive biomarkers in a CPG in 2013 [13].
Which Mutations Should be Tested?
Since close to 90 % of the mutations in lung adenocarcinomas have deletions in exon 19 or point mutation in exon 21 (L858R) of EGFR gene, most laboratories offer molecular tests designed to detect these alterations. However, after the publication of EGFR mutations and ALK rearrangements testing guidelines for selecting patients for TKI therapy, the current recommendation for clinical EGFR mutation testing includes ability to detect all individual mutations that have been reported with a frequency of at least 1 % of EGFR-mutated lung adenocarcinomas (Table 8.3 from guidelines). This is not a concern for laboratories offering Sanger sequencing, which provides information on mutations sequenced throughout the exons 18–21. But it is an important consideration in the selection or design of mutation-specific assays.
Table 8.3
EGFR mutations recommended to be tested for selecting patients for EGFR-TKI therapy
EGFR exon | EGFR codon | Mutations (amino acid) | Nucleotide substitutions | Approximate % of all EGFR mutations |
|---|---|---|---|---|
18 | E709 | E709K E709A E709G E709V E709D E709Q | c.2125G>A c.2126A>C c.2126A>G c.2126A>T c.2127A>C, c.2127A>T c.2125G>C | 1 |
G719 | G719S G719A G719C G719D | c.2155G>A c.2156G>C c.2155G>T c.2156G>A | 2–5 | |
19 | K739 I740 P741 V742 A743 I744 | Insertions 18 bp ins | 1 | |
E746 L747 R748 E749 A750 T751 S752 P753 | Deletions 15 bp del 18 bp del 9 bp del 24 bp del 12 bp del | 45 | ||
20 | S768 V769 D770 N771 P772 H773 V774 | Insertions | 4–10 | |
S768 | S768I | c.2303G>T | 1–2 | |
T790 | T790M | c.2369C>T | 2 | |
21 | L858 | L858R L858M | c.2573T>G c.2572C>A (rare) | 40 |
L861 | L861Q L861R | c.2582T>A, c.2582T>G | 2–5 |
When to Test
EGFR molecular testing should be used to select patients for EGFR-targeted TKI therapy. Patients with lung adenocarcinoma should not be excluded from testing on the basis of clinical characteristics. All patients with advanced lung adenocarcinomas at the time of diagnosis irrespective of morphology and grade and/or at the time of recurrence or progression in patients who originally presented with lower stage disease but were not previously tested are eligible for molecular testing. Lower stage tumors may be tested for mutations but the decision to do so should be made locally by each laboratory, in collaboration with its oncology team. Current summary of guideline recommendation is listed in Table 8.4 [13].
Section I: When should molecular testing of lung cancers be performed? |
Question 1: Which patients should be tested for EGFR mutations and ALK rearrangements? |
1.1a: Recommendation: EGFR molecular testing should be used to select patients for EGFR-targeted TKI therapy, and patients with lung adenocarcinoma should not be excluded from testing on the basis of clinical characteristics |
1.1b: Recommendation: ALK molecular testing should be used to select patients for ALK-targeted TKI therapy, and patients with lung adenocarcinoma should not be excluded from testing on the basis of clinical characteristics |
1.2: Recommendation: In the setting of lung cancer resection specimens, EGFR and ALK testing is recommended for adenocarcinomas and mixed lung cancers with an adenocarcinoma component, regardless of histologic grade. In the setting of fully excised lung cancer specimens, EGFR and ALK testing is not recommended in lung cancers that lack any adenocarcinoma component, such as pure squamous cell carcinomas, pure small-cell carcinomas, or large-cell carcinomas lacking any immunohistochemistry (IHC) evidence of adenocarcinoma differentiation |
1.3: Recommendation: In the setting of more limited lung cancer specimens (biopsies, cytology) where an adenocarcinoma component cannot be completely excluded, EGFR and ALK testing may be performed in cases showing squamous or small-cell histology but clinical criteria (e.g., young age, lack of smoking history) may be useful in selecting a subset of these samples for testing |
1.4: Recommendation: To determine EGFR and ALK status for initial treatment selection, primary tumors or metastatic lesions are equally suitable for testing |
1.5: Expert consensus opinion: For patients with multiple, apparently separate, primary lung adenocarcinomas, each tumor may be tested but testing of multiple different areas within a single tumor is not necessary |
Question 2: When should a patient specimen be tested for EGFR mutation or ALK rearrangement? |
2.1a: Recommendation: EGFR mutation testing should be ordered at the time of diagnosis for patients presenting with advanced-stage disease (stage IV according to the 7th edition TNM staging system) who are suitable for therapy or at the time of recurrence or progression in patients who originally presented with lower stage disease but were not previously tested |
2.1b: Suggestion: ALK rearrangement testing should be ordered at the time of diagnosis for patients presenting with advanced-stage disease (stage IV according to the 7th edition TNM staging system) who are suitable for therapy or at the time of recurrence or progression in patients who originally presented with lower stage disease but were not previously tested |
2.2a: Expert consensus opinion: EGFR testing of tumors at diagnosis from patients presenting with stage I, II, or III disease is encouraged but the decision to do so should be made locally by each laboratory, in collaboration with its oncology team |
2.2b: Expert consensus opinion: ALK testing of tumors at diagnosis from patients presenting with stage I, II, or III disease is encouraged, but the decision to do so should be made locally by each laboratory, in collaboration with its oncology team |
2.3: Recommendation: Tissue should be prioritized for EGFR and ALK testing |
Question 3: How rapidly should test results be available? |
3.1: Expert consensus opinion: EGFR and ALK results should be available within 2 weeks (10 working days) of receiving the specimen in the testing laboratory |
3.2: Expert consensus opinion: Laboratories with average turnaround times beyond 2 weeks need to make available a more rapid test—either in-house or through a reference laboratory—in instances of clinical urgency |
3.3: Expert consensus opinion: Laboratory departments should establish processes to ensure that specimens that have a final histopathologic diagnosis are sent to outside molecular pathology laboratories within 3 working days of receiving requests and to intramural molecular pathology laboratories within 24 h |
Section II: How should EGFR testing be performed? |
Question 4: How should specimens be processed for EGFR mutation testing? |
4.1: Expert consensus opinion: Pathologists should use formalin-fixed, paraffin-embedded (FFPE) specimens or fresh, frozen, or alcohol-fixed specimens for PCR-based EGFR mutation tests. Other tissue treatments (e.g., acidic or heavy metal fixatives, or decalcifying solutions) should be avoided in specimens destined for EGFR testing |
4.2: Expert consensus opinion: Cytologic samples are also suitable for EGFR and ALK testing, with cell blocks being preferred over smear preparations |
Question 5: What are the specimen requirements for EGFR testing? |
5.1: Expert consensus opinion: Pathologists should determine the adequacy of specimens for EGFR testing by assessing cancer cell content and DNA quantity and quality |
5.2: Expert consensus opinion: Each laboratory should establish the minimum proportion and number of cancer cells needed for mutation detection during validation |
5.3: Expert consensus opinion: A pathologist should assess the tumor content of each specimen and either perform or guide a trained technologist to perform microdissection for tumor cell enrichment as needed |
Question 6: How should EGFR testing be performed? |
6.1: Recommendation: Laboratories may use any validated EGFR testing method with sufficient performance characteristics |
6.2: Expert consensus opinion: Laboratories should use EGFR test methods that are able to detect mutations in specimens with at least 50 % cancer cell content, although laboratories are strongly encouraged to use (or have available at an external reference laboratory) more sensitive tests that are able to detect mutations in specimens with as little as 10 % cancer cells |
6.3: Expert consensus opinion: Clinical EGFR mutation testing should be able to detect all individual mutations that have been reported with a frequency of at least 1% of EGFR-mutated lung adenocarcinomas |
6.4: Recommendation: Immunohistochemistry for total EGFR is not recommended for selection of EGFR TKI therapy |
6.5: Recommendation: EGFR copy number analysis (i.e., FISH or CISH) is not recommended for selection of EGFR TKI therapy |
Question 7: What Is the role of KRAS analysis in selecting patients for targeted therapy with EGFR TKIs? |
7.1: Recommendation: KRAS mutation testing is not recommended as a sole determinant of EGFR TKI therapy |
Question 8: What additional testing considerations are important in the setting of secondary or acquired EGFR TKI resistance? |
8.1: Recommendation: If a laboratory performs testing on specimens from patients with acquired resistance to EGFR kinase inhibitors, such tests should be able to detect the secondary EGFR T790M mutation in as few as 5% of cells |
Section III: How should ALK testing be performed? |
Question 9: What methods should be used for ALK testing? |
9.1: Recommendation: Laboratories should use an ALK FISH assay using dual-labeled break-apart probes for selecting patients for ALK TKI therapy; ALK immunohistochemistry, if carefully validated, may be considered as a screening methodology to select specimens for ALK FISH testing |
9.2: Recommendation: RT-PCR is not recommended as an alternative to FISH for selecting patients for ALK inhibitor therapy |
9.3: Expert consensus opinion: A pathologist should be involved in the selection of sections for ALK FISH testing, by assessing tumor architecture, cytology, and specimen quality |
9.4: Expert consensus opinion: A pathologist should participate in the interpretation of ALK FISH slides, either by performing the analysis directly or by reviewing the interpretations of cytogeneticists or technologists with specialized training in solid tumor FISH analysis |
9.5: Expert consensus opinion: Testing for secondary mutations in ALK associated with acquired resistance to ALK inhibitors is not currently required for clinical management |
Section IV: Should other genes be routinely tested in lung adenocarcinoma? |
Question 10: Are other molecular markers suitable for testing in lung cancer? |
10.1a: Recommendation: Testing for EGFR should be prioritized over other molecular markers in lung adenocarcinoma |
10.1b: Suggestion: After EGFR testing, testing for ALK should be prioritized over other proposed molecular markers in lung adenocarcinoma, for which published evidence is insufficient to support testing guideline development at the present time |
Section V: How should molecular testing of lung adenocarcinomas be implemented and operationalized? |
Question 11: Must all adenocarcinomas be tested for both EGFR and ALK? |
11.1: Expert consensus opinion: Laboratories may implement testing algorithms to enhance the efficiency of molecular testing of lung adenocarcinomas, provided the overall turnaround time requirements are met |
Question 12: How should EGFR and ALK results be reported? |
12.1: Expert consensus opinion: EGFR mutation testing reports and ALK FISH reports should include a results and interpretation section readily understandable by oncologists and by nonspecialist pathologists |
Question 13: How should EGFR and ALK testing be validated? |
13.1: Expert consensus opinion: EGFR and ALK testing validation should follow the same guidelines as for other molecular diagnostics and FISH tests |
Question 14: How should quality assurance be maintained? |
14.1: Expert consensus opinion: Laboratories should follow similar quality control and quality assurance policies and procedures for EGFR and ALK testing in lung cancers as for other clinical laboratory assays. In particular, laboratories performing EGFR and ALK testing for TKI therapy should enroll in proficiency testing, if available |
Sample Requirements
The effects of routine histology processing on any molecular diagnostics applications that rely on PCR are not different for EGFR mutation analysis. EGFR testing can be performed on fresh, frozen, FFPE, or alcohol-fixed specimens. The selection of the specimen to be tested should be performed by pathologists who also determine the adequacy of specimens for EGFR testing by assessing cancer cell content and DNA quantity and quality. Tumor content may be enriched by directing technologists to microdissect the tumor. Every CLIA-certified laboratory offering molecular tests should validate all types of specimen types and processing variations likely to be encountered (i.e., FFPE, fresh, frozen, alcohol fixed), and testing should be performed and reported only on validated specimen types. Other specimen types including tumor samples with inadequate tumor content should be rejected as inadequate or tested at the discretion of the laboratory director, with clear communication on the report that the test was performed on a nonvalidated specimen type.
Brief Overview of Common Platforms Currently Used for EGFR Testing
Laboratories may use any testing method that is validated for EGFR mutation detection with sufficient performance characteristics. These include ability to detect mutations in specimens with at least 50 % cancer cell content, but are encouraged to be able to detect mutations in specimens with as little as 10 % cancer cells. Different methodologies exist in the published literature, each having different advantages and disadvantages. They include Sanger sequencing with and without mutated allele enrichment strategies [e.g., locked nucleic acid (LNA), peptide nucleic acid clamps (PNA)], the amplification refractory mutation system (ARMS-PCR), length analysis, restriction fragment length polymorphism, real-time PCR, high-resolution melting curve analysis, single-base extension genotyping (including mass spectrometry-based genotyping), denaturing high-performance liquid chromatography, and more recently next-generation sequencing platforms. Common testing methodologies are described below.
Length Analysis and Restriction Fragment Length Polymorphism
Deletions in exon 19 of EGFR : Most commonly used assay to detect deletions in exon 19 of EGFR is length analysis of fluorescently labeled PCR products. If there is presence of mutation, the mutant allele is of smaller size than the wild type and therefore can be easily detected by capillary electrophoresis (Fig. 8.1). Different size deletions can be discriminated from wild-type allele by the difference in size of the length of PCR product.
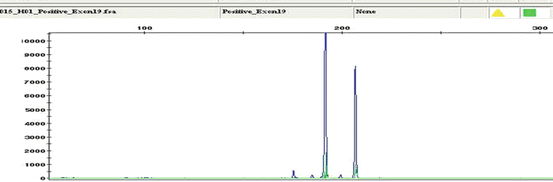

Fig. 8.1
EGFR exon 19 deletion (15 bp deletion) mutant peak being the shorter allele
EGFR exon 21 point mutation: The 2573 T>G (L858R) mutation creates a new Sau96I restriction site, GGNCC. Incubation and digestion of PCR product with restriction enzyme Sau96I generate an additional shorter fragment, the basis for a PCR restriction fragment length polymorphism (PCR-RFLP) assay design. The digested fluorescently labeled PCR products are then analyzed by capillary electrophoresis [32] (Fig. 8.2). The sensitivity or limit of detection for both the assays is estimated between 3 and 6 %.
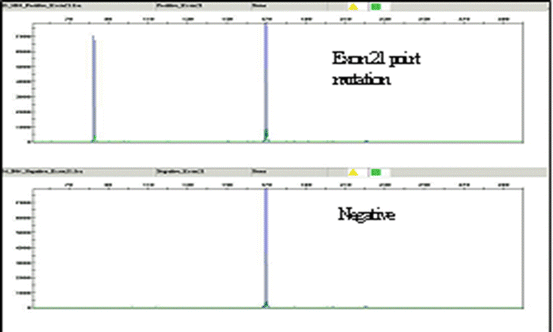

Fig. 8.2
EGFR exon 21 point mutation (L858R) with PCR-RFLP-based assay
Amplification Refractory Mutation System-PCR
ARMS-PCR uses the ability of Taq DNA polymerase effectively distinguishing between a match and a mismatch at the 3′ end of a PCR primer. Specific mutated sequences are selectively amplified with maximum efficiency only when the primer is fully matched, in a background of the sequences that do not carry the mutation including wild-type sequences. Only a low-level background amplification occurs when the 3′ base is mismatched. The fluorescently labeled probes are called scorpions which have fluorophore and quencher in close proximity leading to reduced fluorescence [fluorescence resonance energy transfer (FRET)]. Upon successful PCR, the fluorophore separates leading to increased fluorescence. A commercially available assay detecting 29 mutations in EGFR gene in exons 18–21 is available using this method (Fig. 8.3). The sensitivity for all the mutations except T790M is about 1 %. For T790M, the sensitivity is about 10 %.
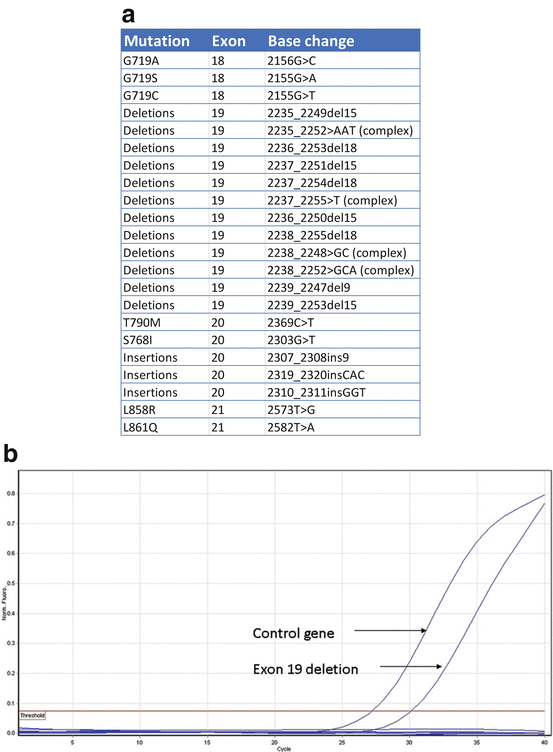

Fig. 8.3
(a) Qiagen RGQ mutation assay: designed to detect 28 mutations associated with TKI sensitivity and one resistance mutation in exon 20 (T790M). (b) ARMS PCR plots showing examples of exon 19 mutation
Sanger Sequencing (SS)
Direct Sanger sequencing has been the traditional method to detect mutations in different genes including EGFR exons 18–21. It is generally accepted that direct sequencing is likely to miss mutations when the tumor cell content is less than 25 % of the sample. Therefore, although a gold standard, SS may not be sensitive enough to detect mutations in a limited tissue sample such as needle biopsies or fine-needle aspiration cytology material. In a limited sample, enrichment strategies for mutant allele using LNA or PNA probe may be necessary for acceptable clinical sensitivity. An example of deletion in exon 19 of EGFR is shown in Fig. 8.4.
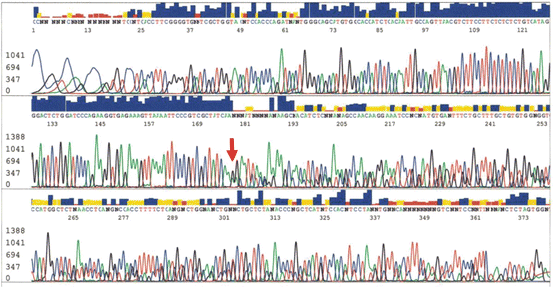

Fig. 8.4
EGFR exon 19 deletion mutation: arrow showing boundary of the deletion displaying overlapping sequences of the shorter allele
Mass Spectrometry: Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) MassARRAY system (Sequenom), originally designed to analyze single-nucleotide polymorphisms in amplified DNA fragments, is also used to detect known somatic point mutations [33]. Genotyping is done by single-allele base extension reaction products that discriminate mutant and wild-type alleles for a given point mutation based on mass differences that are specific to the sequence of the wild-type product and the mutant products as resolved by MALDI-TOF MS [34].
The sensitivity of detection of mutation is generally accepted between 1 and 10 % depending upon plex level of the assay. Example of positive exon 21 L858R mutation is shown in Fig. 8.5. The wild-type and mutant peaks are discriminated based on differences in the masses of the single-base extension with mass-modified dNTPs.
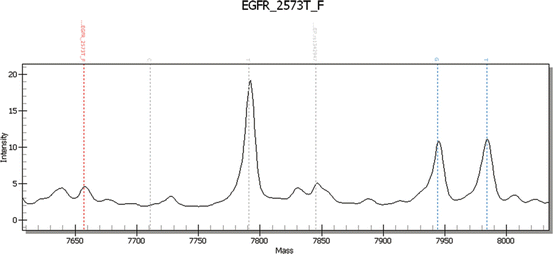

Fig. 8.5
EGFR exon 21 2573 T>G (L858R) wild-type and mutant alleles highlighted in blue dotted lines. Courtesy: Laetitia Borsu, Ph.D. Assistant Attending, Genomics Project Manager, MSKCC, New York, NY
Significance of Negative Results
Negative results for EGFR indicate lack of clinically significant response to EGFR TKI therapy.
Is the Test Adaptable to Immunohistochemistry?
Immunohistochemistry (IHC) for total EGFR is not recommended for selection of EGFR TKI therapy due to poor correlation between total EGFR expression and mutation status. In addition, response to EGFR TKI does not correlate with total EGFR expression detected by using immunohistochemistry [19, 35, 36]. IHC for phosphorylated EGFR is limited as the stability of phosphorylation status in FFPE material is variable. EGFR mutation-specific monoclonal antibodies against mutant protein, viz. L858R point mutation in exon 21 and 15 bp deletion in exon 19 (most common deletion), have been shown to be sensitive and specific to detect these mutations in the published literature [37–39]. But for exon 19 deletions, the sensitivity is reduced to detect other sized deletions. These mutation-specific antibodies could be used to identify patients eligible for EGFR TKI therapy. However, mutation-specific IHC is very insensitive to be used as a stand-alone assay. Also, for negative results, currently there is no algorithm that exists to test for mutations and molecular testing is still needed. In addition, no prospective clinical experience is available for validation. IHC may be an option in small low-cellularity diagnostic specimens with marginal tumor cellularity for DNA-based analysis, but again negative results do not replace mutation testing [40]. Per published guidelines for EGFR testing for selecting patients eligible for EGFR TKI, the body of published data is insufficient to make an evidence-based recommendation regarding the use of EGFR mutation-specific IHC at this time [13].
FISH or CISH Testing for EGFR Amplification
EGFR amplification or polysomy is frequently encountered in patients with lung adenocarcinomas but the response rate with EGFR TKI for these patients is well below the response rate in patients with EGFR mutations (30 vs. 68 %) [35, 36]. Amplification of mutant allele is noted commonly [19] and that is strongly associated with EGFR polysomy/amplification. This association is thought to be related to EGFR TKI response. But in cases where there is discrepancy between EGFR mutation and copy number, the mutation status is better predictor response to EGFR TKI [36]. Therefore, EGFR copy number analysis (FISH or chromogenic in situ hybridization) is not recommended for selection of EGFR TKI therapy [13].
ALK (Anaplastic Lymphoma Receptor Tyrosine Kinase)
ALK gene, located on chromosome 2p23, encodes a receptor tyrosine kinase, which belongs to the insulin receptor superfamily [41]. ALK plays an important role in the development of the brain and exerts its effects on specific neurons in the nervous system. This gene is frequently rearranged with different genes with novel fusions, mutated, or amplified in many tumors including anaplastic large-cell lymphomas, neuroblastoma, and NSCLC [41].
ALK Mutations
In 2007, Soda and co-workers reported an interstitial deletion and inversion within the short arm of chromosome arm 2 that resulted in the formation of a novel fusion gene comprising portions of the echinoderm microtubule-associated protein-like 4 (EML4) gene and the ALK gene in a subset of lung adenocarcinoma [42]. The chimeric fusion of EML4-ALK results in constitutive dimerization leading to oncogenic activity of ALK gene [43]. The original report from Soda et al. showed 7 % positive rate in NSCLC in Japanese patients (5 of 75). Subsequent studies in the USA have shown the gene fusion positive rate in about 2–7 % of all NSCLCs. This gene fusion is more frequent in adenocarcinomas in never smokers or light smokers.
Patients with tumors containing EML4-ALK mutation respond effectively with ALK kinase inhibitors alone or in combination [44, 45]. In a large series ALK rearrangement-positive (about 5 % of 1,500 NSCLC patients screened) patients treated with EGFR inhibitor, crizotinib, showed an overall response rate of 57 %, with 72 % having a PFS of 6 months or greater [46]. The US Food and Drug Administration (FDA) approved crizotinib for advanced-stage, ALK-positive lung cancer in November 2013. This targeted therapy is also recommended by guidelines from professional organizations, including the American Society of Clinical Oncology (ASCO), European Society for Medical Oncology, and National Comprehensive Cancer Network (NCCN). To that effect as described above CAP/IASLC/AMP published evidence-based recommendations for the molecular testing of lung cancers for ALK as second predictive biomarkers in a CPG in 2013 [13]
Molecular Testing for ALK Mutations
ALK gene rearrangement can be detected using RT-PCR, direct Sanger sequencing, fluorescence in situ hybridization (FISH), and immunohistochemistry.
RT-PCR
To detect all fusion products, RT-PCR has limitations as there are many fusion combinations between different exons on EML4 and ALK genes and each needs a separate primer pair. Also, rare other fusion partners with ALK (KIF5B–ALK, TFG–ALK [47, 48]) will be missed by RT-PCR as the primers are designed for specific breakpoints in different exons [49]. Thus there are concerns for a higher failure rate of an RNA-based assay in routine FFPE pathology material and RT-PCR is currently not recommended as a first-line diagnostic method for determining ALK fusion status
Immunohistochemistry
IHC is an attractive and cheaper way to detect ALK overexpression as most anatomic pathology departments have IHC laboratory and pathologists use IHC extensively in their routine practise. However, due to low expression of ALK protein in ALK-rearranged lung adenocarcinomas, IHC is not reliable using the same antibody clone compared to anaplastic large-cell lymphoma (mouse monoclonal anti-human CD246, clone ALK1). Recently two rabbit monoclonal anti-human ALK antibodies (clones D5F3 and D9E4) have shown high sensitivity, specificity, and reproducibility compared to ALK FISH. These reports show promising results and IHC assays may potentially facilitate the routine identification of ALK-rearranged lung adenocarcinoma [50]. The published view by CAP/IASLC/AMP based on current literature is that the data is still limited and more studies are needed to recommend developing a specific recommendation on the use of ALK IHC as a sole determinant of ALK TKI therapy [13, 124].
ALK FISH
The commercial availability of a dual-probe “break-apart” (FISH) assay for ALK fusions/rearrangements is the most efficient and consistent way to detect ALK gene rearrangements for clinical use. This is the same ALK FISH dual probe used to detect anaplastic large-cell lymphoma [51]. This commercially available ALK FISH assay is FDA approved to be used as a companion diagnostic assay for selecting patients for crizotinib therapy.
Specimen Requirements
The consensus for specimen adequacy for FISH testing is similar to EGFR testing (see above) and other FISH assays used in molecular genetic laboratories using FFPE tissue sections. Each laboratory is required to validate the FISH assay on internal samples which are expected in routine testing before offering the test for clinical use. These include different fixatives (FFPE, alcohol) and specimen types (surgical biopsy, cytology specimens, etc.). DNA-damaging fixatives, decalcified tissues in general, are not suitable for any molecular assay. A pathologist should assess the adequacy of the tumor proportion in a given specimen and mark the most cellular area for scoring. The minimum tumor cell content/requirement, although similar to EGFR testing, is less stringent as FISH is a direct visualization and scoring assay. Interpretations should be done by a trained pathologist either by directly screening and scoring the FISH slide or by reviewing the results of cytogeneticists or technologist with specialized training in solid tumor FISH analysis.
Assay Interpretation
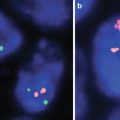
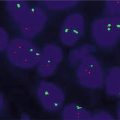
Negative results have a yellow fusion signal or narrowly split orange/red and green signals using break-apart dual-labeled FISH assay (Fig. 8.6). The most common positive result in lung cancer will result in one separate orange/red and one separate green signal (Fig. 8.7). The separation of orange/red and green signals should be a gap larger than two signal diameters, to be indicative of an ALK gene rearrangement. The germline unaltered ALK region servers as a built-in negative control which stays as a yellow fusion signal or appears as two narrowly split orange/red and green signals. The second positive result is loss of the green 5′ probe with a remaining unpaired 3′ orange/red probe, indicating an unbalanced rearrangement (Fig. 8.8). Minimum of 50 tumor nuclei in a tumor-rich area are recommended for adequate scoring. The tumors are considered positive if 15 % or more of 50 nuclei assessed show the classic split-signal pattern or loss of the green 5′ probe with remaining red 3′ probe [51, 52]. This cutoff is part of the labeling of the FDA-approved commercial FISH assay.
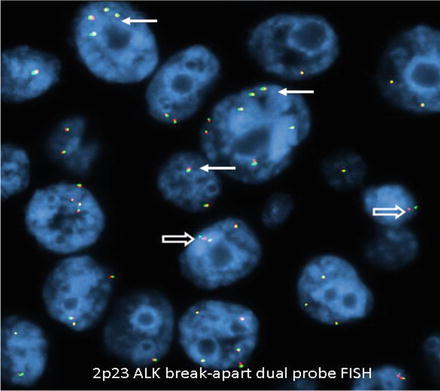
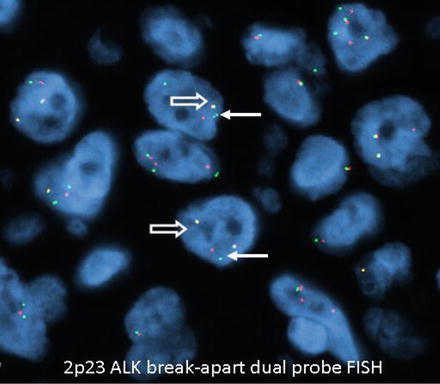
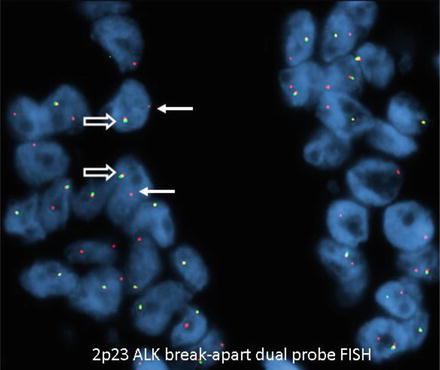

Fig. 8.6
Negative for ALK rearrangement. Arrows: Fusion yellow signal. Open arrows: Narrowly split orange/red and green signals interpreted as negative for ALK rearrangement. Note aneuploidy of chromosome 2p (original magnification × 1,000)

Fig. 8.7
Positive for ALK rearrangement. Arrows: One separate orange/red and one separate green signal. Open arrows: Fusion yellow signal built in negative control (wild type) (original magnification × 1,000)

Fig. 8.8
Positive for ALK rearrangement (single 3′ ALK). Arrows: Loss of the green 5′ probe with remaining red 3′ probe. Open arrows: Fusion signal serving as built in negative control (wild type) (original magnification × 1,000)
Other Gene Mutations That Alter Response to Current Targeted Therapy With EGFR inhibitors or Show Potential For Targeted Therapy in Lung Cancer
KRAS (Kirsten Rat Sarcoma Viral Oncogene Homolog)
KRAS gene, located on chromosome 12p12.1, encodes a protein which is member of the small GTPase superfamily [53]. Ras proteins bind GDP/GTP and possess intrinsic GTPase activity. In many malignancies a single amino acid substitution leads to an activating mutation that results in constitutive activation of the protein leading up-regulation of downstream signaling cascade including MAPK kinase, PI3 kinase signaling [53], etc.
KRAS Mutations
KRAS mutations are seen in 15–25 % of lung adenocarcinomas and are uncommon in squamous cell carcinomas (SCC) [54]. Most mutations are point mutations producing an amino acid substitution at codons 12, 13, or 61 leading to constitutive activation of downstream signaling pathways. KRAS, EGFR, and ALK mutations are mutually exclusive in NSCLC. KRAS mutations are more frequently encountered in adenocarcinoma with mucinous morphology compared to other morphologies and in patients with history of smoking, both former/current smokers and never smokers [55, 56]. Five percent of never smokers with lung adenocarcinoma have been shown to harbor KRAS mutations [57]. In contrast to colorectal adenocarcinomas, in advanced NSCLC, the role of KRAS as a biomarker either as a prognostic or predictive marker is uncertain at this time as very few prospective randomized trials have been completed [55, 58].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree