Liquid Biopsies in Gastrointestinal Cancers
Shalini Makawita
Arvind Dasari
Introduction
In this section we will discuss liquid biopsies, specifically pertaining to cell-free DNA (cfDNA) or circulating tumor DNA (ctDNA), and its applicability in the management of gastrointestinal (GI) cancer patients.
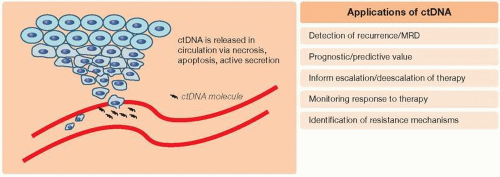
Liquid biopsies are simple, noninvasive tests in biologic fluids such as blood or urine that enable the measurement of clinically relevant biomarkers. There are many analytes within the circulatory system that can serve as markers of disease states, such as proteins, messenger mRNA, micro RNA (miRNA), and cell-free DNA (cfDNA). cfDNA had been first reported in 1948 by Mandel and Metais in human plasma and consists of fragments of genetic material found outside of the cell (1). In 1977, Leon et al. (2) had reported elevated levels of cfDNA in cancer patients when compared to healthy individuals. cfDNA that has been released from tumor cells is referred to as ctDNA and can vary in range from a minute portion (<1%) of an individual’s total cfDNA to a relatively high abundance correlating with tumor burden. cfDNA or ctDNA can serve as a surrogate marker of disease in cancer patients, and three potential mechanisms for its release into circulation have been described: necrosis, apoptosis, and active secretion (Figure 10.1) (3).
Over the past decades, with significant advancements in methods for ctDNA detection, its applicability in the clinical care of cancer patients has been garnering significant interest and research. This includes its use as prognostic and/or predictive markers, in the detection of disease recurrence after definitive therapy, in the management of minimal residual disease (MRD), in monitoring response to therapy, and in identification of resistance mechanisms after exposure to targeted therapies (Figure 10.1). Through sampling of the blood for ctDNA at multiple time points during a patient’s cancer treatment, liquid biopsies can aid in the longitudinal evaluation of a patient’s cancer genome. In the sections below, we detail clinical applications of ctDNA within the context of common GI malignancies.
Colorectal Cancer
Several early studies by multiple groups had shown individual cfDNA markers or marker panels as well as methylation patterns to have the ability to distinguish patients with colorectal cancer (CRC) from benign or healthy controls. These early studies had also established good concordance with tissue molecular profiling (4,5). In a study where fresh frozen tissue and plasma samples obtained at the time of surgery from 29 CRC patients were analyzed with an 85-gene panel, median ctDNA levels of 14.2 and 8.94 ng/mL were noted in colon and rectal cancer patients, respectively, with 70% concordance between ctDNA and tissue molecular profile. The small number of rectal cancer patients in this study (N = 10) had lower overall concordance (50%) with tissue profiles (6). Systematic reviews of literature published over the past two decades have noted other individual ctDNA markers such as septin 9 (SEPT9), or marker panels, that have been studied for disease detection. Varying ranges of sensitivity (37%-96%) and specificity (79%-99%) among studies have excluded these markers as clinically useful screening tools but with potential for other applications such as disease monitoring with further validation (7). Methylation status of promoter regions of genes methylguanine methyltransferase (MGMT) and DNA excision repair protein (ERCC-1) analyzed via liquid biopsy in 50 patients with rectal cancer compared to 43 patients with benign rectal lesions had also shown discriminatory potential with a specificity of 93% to 95% and sensitivity of approximately 60% (8). Petit et al. (7) from their systematic review concluded that methylated cfDNA may serve as a promising candidate for CRC detection.
With the advent of next-generation sequencing (NGS), genomic profiling using various commercially available liquid biopsy platforms has enabled serial analysis of ctDNA for prediction of survival outcomes and detection of disease recurrence. MRD is defined as minute amounts of cancer cells remaining during or after completion of therapy as detected by highly sensitive techniques. MRD can imply the presence of micrometastasis prior to clinical detection. Identification of ctDNA after definitive surgery in colon cancer patients has been associated with increased disease recurrence. In a study of 231 stage II colon cancer patients, ctDNA had been detected in 14/178 patients postoperatively who did not receive adjuvant therapy, and 11/14 (79%) patients had recurrence of disease at 27 months follow-up compared to a recurrence rate of 16/164 (9.8%) patients who did not have ctDNA present postoperatively (hazard ratio [HR] 18; 95% confidence interval [CI] 7.9-40;
p < 0.001) (9). In patients who received adjuvant chemotherapy, ctDNA positivity directly after completion was associated with worse recurrence-free survival (RFS) (HR 11; 95% CI 1.8-68; p = 0.001). Reinert et al. (10) used ultradeep multiplex polymerase chain reaction (PCR)-based NGS sequencing of ctDNA in their prospective multicenter cohort study of 130 patients with stage I-III CRC at selected time points, including before surgery, on postoperative day 30, and once every 3 months for up to 3 years to evaluate its utility in detecting clinical recurrence and RFS. Using WES of tumor to identify 16 highly ranked somatic single-nucleotide variants and short indels, this group used a technology to construct personalized liquid biopsies for each patient. Preoperatively, 88.5% of patients had detectable ctDNA. Post-op ctDNA positivity was associated with seven times more likelihood of relapse (HR 7.2; 95% CI 2.7-19.0; p < 0.001), and ctDNA positivity after adjuvant chemotherapy was associated with 17 times more likelihood of relapse (HR 17.5; 95% CI 5.4-56.5; p < 0.001). Currently, although such assays are predictive of survival outcomes, prospective clinical trials are needed to determine if intervention with additional therapy based on ctDNA detection will improve these outcomes.
p < 0.001) (9). In patients who received adjuvant chemotherapy, ctDNA positivity directly after completion was associated with worse recurrence-free survival (RFS) (HR 11; 95% CI 1.8-68; p = 0.001). Reinert et al. (10) used ultradeep multiplex polymerase chain reaction (PCR)-based NGS sequencing of ctDNA in their prospective multicenter cohort study of 130 patients with stage I-III CRC at selected time points, including before surgery, on postoperative day 30, and once every 3 months for up to 3 years to evaluate its utility in detecting clinical recurrence and RFS. Using WES of tumor to identify 16 highly ranked somatic single-nucleotide variants and short indels, this group used a technology to construct personalized liquid biopsies for each patient. Preoperatively, 88.5% of patients had detectable ctDNA. Post-op ctDNA positivity was associated with seven times more likelihood of relapse (HR 7.2; 95% CI 2.7-19.0; p < 0.001), and ctDNA positivity after adjuvant chemotherapy was associated with 17 times more likelihood of relapse (HR 17.5; 95% CI 5.4-56.5; p < 0.001). Currently, although such assays are predictive of survival outcomes, prospective clinical trials are needed to determine if intervention with additional therapy based on ctDNA detection will improve these outcomes.
Currently, the standard of care is to offer adjuvant chemotherapy after surgical resection for patients with stage III colon adenocarcinoma and high-risk stage II disease (T4, inadequate lymph node harvest [≤12], tumors complicated by obstruction or perforation, poorly differentiated or vascular/perineural or lymphatic invasion) (11). There are no validated biomarkers in standard clinical practice for identification of MRD after surgical resection in colon cancer; however, ongoing prospective studies incorporating ctDNA analyses are underway to inform clinical decision-making with regard to adjuvant therapy. In the prospective phase II/III COBRA trial (NCT04068103), patients with stage IIA disease without high-risk features after surgical resection who are deemed candidates for observation and no systemic chemotherapy will be randomized 1:1 to observation or prospective testing of ctDNA for both colon cancer-relevant genetic mutations and methylation profiling (12). Those in the ctDNA arm with detectable ctDNA will be treated for 6 months with adjuvant 5-fluorouracil, leucovorin, oxaliplatin (FOLFOX)/capecitabine, oxaliplatin (CAPOX) chemotherapy. Primary endpoints of this study include the clearance of ctDNA with use of adjuvant systemic therapy and RFS in patients with detected ctDNA treated with or without systemic therapy.
Other clinical trials exploring the utility of ctDNA for informing adjuvant chemotherapy include the Australian DYNAMIC III study (ACTRN12617001566325) in stage III colon cancer, and the CIRCULATE-Japan and US trials (NCT04120701) (13). Data from the observational GALAXY study within CIRCULATE-Japan evaluating a total of 1,365 patients with stage I-IV CRC showed improved 6-month disease-free survival (DFS) rate in patients with positive ctDNA at 4 weeks who turned negative at 12 weeks post-op (N = 58) compared to those whose ctDNA remained positive at 4 and 12 weeks (N = 78) (DFS 100% vs. 45%; HR 52.3; p < 0.001). Clearance of ctDNA at 12 weeks was significantly higher in those who received adjuvant chemotherapy (57% vs. 8%, all stages p < 0.001), and 6-month DFS rate was significantly higher in ctDNA-positive patients who received adjuvant chemotherapy (84% vs. 34%; HR 0.15; p < 0.001) (14,15). ctDNA informed therapy escalation/deescalation is ongoing. These studies are summarized in Table 10.1.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree