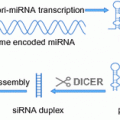
Fig. 1
a Schematic overview of HPV infection and integration during the development of HPV-positive tumors (modified from Woodman et al. 2007; Cornet et al. 2015). HPV is thought to access the basal cells through micro lesions in the squamous cell epithelium. Following infection, the early HPV genes E1, E2, E4, E5, E6 and E7 are expressed and the viral DNA replicates from episomal DNA. In the upper layers of the epithelium, the viral genome is replicated further, and the late genes L1 and L2, and E4 are expressed. L1 and L2 allow encapsulation of the viral genomes to form progeny virions in the nucleus. The shed virus can then initiate a new infection. In the transition to (micro)invasive cancer, viral DNA often integrates in 1 or more copies into the host genomic DNA, with often associated loss or disruption of E2, and subsequent upregulation of E6 and E7 oncogene expression. LCR, long control region. b The subsequent upregulation of E6 and E7 oncoproteins results in deregulation of cell signaling pathways, which, among others, leads to increased cellular proliferation and inhibition of apoptosis (modified from Olthof et al. 2012; Groves and Coleman 2015). c Multiple mechanisms by which HPV integration into the host genome may directly lead to deregulation of the key cellular tumor suppressor genes and proto-oncogenes (modified from Rusan et al. 2015)
2 Mechanisms Involved in and Approaches to Detect HPV Integration
Persistent infection may also result in integration of the HRHPV genome or parts thereof into the host genome. Although in premalignant CIN lesions the time and frequency of integration has been heavily debated, it is now believed that it occurs relatively late in the progression of high-grade dysplasia to (micro)invasive anogenital carcinomas (Klaes et al. 1999; Hopman et al. 2004; Vinokurova et al. 2008; Rusan et al. 2015). HPV16 integration could also be detected in OPSCC and in tumor adjacent dysplasia in some of these cases (Hafkamp et al. 2003; Mooren et al. 2014). So far, in situ localization of a persistent HRHPV infection in the oropharynx/palatine tonsils has been extremely difficult to find and detect in the normal population (Klingenberg et al. 2010), and analysis of tissue biopsies of a population with a high chance of HPV infection (e.g., people having many sex partners, oral sex or who are immunosuppressed) is probably required to successfully identify such infections. In contrast, LRHPV infections are easy to detect, for example in laryngeal papillomas, but in these cases viral integration is a seldom finding (Huebbers et al. 2013; Mooren et al. 2014).
Because viral integration requires the breakage of both the viral and the host DNA, the integration rate is believed to be linked to the levels of DNA damage (Chen et al. 2014). DNA damage can be caused by both endogenous and exogenous factors, including inflammation induced either by the virus itself (E6 and E7 expression) or by co-infections with other agents (both resulting in production of excessive amounts of reactive oxygen and nitrate species), environmental agents and other factors (Wei et al. 2009; Lace et al. 2015; Visalli et al. 2016). In this respect, the activation of DNA damage repair mechanisms as well as the accumulation of chromosomal alterations may also contribute to the viral integration process (Southern et al. 2001; Hopman et al. 2004, 2006).
In uterine cervical squamous cell carcinomas (UCSCC), which are HPV positive in 95–100 % of cases, different HRHPV types tend to integrate at different frequencies, such as HPV16 at 50–80 %, HPV18 at >90 %, HPV31 and −33 at 15–40 % and HPV45 at > 80 % (Wentsenzen et al. 2004; Vinokurova et al. 2008; Olthof et al. 2012; Groves and Coleman 2015). In OPSCC HPV, positivity range from 20 to 90 % in different studies and depend among others on the geographical location, sample preparation and detection methods used (Olthof et al. 2012). 90–95 % of virus-positive OPSCC are infected with HPV16, and integration percentages range between 40 and 80 % dependent on the methods used to identify integrated HPV.
Along with the many HPV detection methods available to date (Snijders et al. 2010), a number of approaches have been developed to specifically detect integrated HPV. On the one hand, approaches have been designed to only identify integration events that are transcriptionally active detecting virus–host fusion transcripts, such as RNA in situ hybridization (ISH) (Van Tine et al. 2004b), 3′ RACE–PCR [also known as “Amplification of Papillomavirus Oncogene Transcripts” (APOT) PCR] (Klaes et al. 1999; Lace et al. 2011; Olthof et al. 2014, 2015; Vojtechova et al. 2016) and RNASeq (Akagi et al. 2014; Ojesina et al. 2014; Parfenov et al. 2014; Hu et al. 2015). On the other hand, procedures have been used to detect integrated HPV genomes (regardless of their transcriptional activity), including DNA (F)ISH (Cooper et al. 1991; Hopman et al. 2004; Hafkamp et al. 2008), Southern blotting (Cullen et al. 1991; Cooper et al. 1991; Vojtechova et al. 2016), detection of integrated papillomavirus sequences (DIPS) PCR (Luft et al. 2001; Peter et al. 2010; Huebbers et al. 2013; Li et al. 2013; Olthof et al. 2014, 2015), restriction-site PCR (Thorland et al. 2000), quantitative PCR (Peitsaro et al. 2002; Nagao et al. 2002; Ziegert et al. 2003) and DNASeq (Xu et al. 2013; Akagi et al. 2014; Parfenov et al. 2014, Chandrani et al. 2015; Hu et al. 2015). These analyses have contributed significantly to our current knowledge on the frequency of HPV integration in UCSCC and OPSCC and its impact on cancer development and progression as well as on viral (onco)gene and human gene expression. However, all these assays also have their (dis)advantages and differ in their detection sensitivities, which have to be taken into account when comparing reported data and generating general conclusions on these issues (below).
3 Identification of HPV Integration Sites in the Human Genome
Identification of sites in the human cellular genome where HPV integration events occur is a longstanding field of interest in HPV research. Molecular studies have provided evidence that often 1 and sometimes >1 integration site(s) can be detected in UCSCC and OPSCC (Hopman et al. 2004; Hafkamp et al. 2008; Peter et al. 2010; Mooren et al. 2013; Akagi et al. 2014; Ojesina et al. 2014; Parfenov et al. 2014; Hu et al. 2015). HPV integration sites appear to be distributed all over the human genome in both UCSCC and OPSCC, and lie often within, or close to, fragile sites (Wentsenzen et al. 2004; Akagi et al. 2014; Ojesina et al. 2014; Olthof et al. 2014, 2015; Parfenov et al. 2014; Hu et al. 2015). Furthermore, a number of cytogenetic bands have been identified as integration hotspots, including 3q28, 4q13.3, 8q24.21, 13q22.1 and 17q21.2 accounting for integration sites of >20 % of UCSCC analyzed (Schmitz et al. 2012; Olthof et al. 2014; Chandrani et al. 2015). In addition, Parfenov et al. (2014) and Hu et al. (2015) reported that integration in both UCSCC and OPSCC is often in regions of microhomology (1–10 bp) among the viral and host genome, indicating that fusion between viral and human DNA may have occurred by microhomology-mediated DNA repair pathways. Most frequently integration is detected into genic regions and to a lesser extent in miRNA regions. Parfenov et al. (2014) reported that in 54 % of OPSCC HPV integrated into a known gene (e.g., RAD51B), and in 17 % within 20 kb of a gene. Similarly, Olthof et al. identified in 29 OPSCC 37 HPV16 integration sites, 27 of which were in known or predicted genes, including 17 with a known role in tumorigenesis, such as BCL2, FANCC, HDAC2 and TP63. Hu et al. (2015) reported integration hot spots (range 4.9–9.7 %) in POU5F1B, FHIT, KLF12, KLF5, LRP1B, LEPREL1, HMGA2, DLG2 and SEMA3D, whereas Ojesina et al. (2014) found virus breakpoints in MYC, ERBB2, TP63, FANCC, RAD51B and CEACAM5, both in UCSCC. Also in 7 often used HPV16-positive HNSCC cell lines 2–7 integration sites per nucleus were identified, with integration in genes (DIAPH2, TP63, C9orf156) and intergenic regions (Olthof et al. 2014). Akagi et al. (2014) were able to confirm these observations in cell lines as well as primary tumor specimens and, moreover, found that sites of integration cluster near sites of structural alterations (amplifications, deletions) in the genome. These findings have also been described previously for UCSCC (Lockwood et al. 2007; Peter et al. 2010; Ojesina et al. 2014). As a result, Akagi et al. (2014) proposed a viral genome looping model to explain HPV-driven amplifications and rearrangements that occur at sites of integration, which may be further propagated throughout the genome. It consists of the following steps: (1) host genome and viral episome are nicked, (2) linear HPV genome integrates in cellular genome, (3) circular DNA containing both host and viral sequences is formed, (4) this template is amplified by rolling circle amplification and (5) integrated concatemers of viral–host sequences are generated that might spread further in the genome. Indeed, in the HPV16-positive HNSCC cell lines described by Olthof et al. (2015), FISH experiments provided evidence for multiplication and translocation events of chromosomes harboring integrated viral DNA sequences as well as genomic instability. It should be noted, however, that the looping model is particularly based on analysis of tumor cell lines, which might also have accumulate additional chromosomal alterations induced by long-term cultivation. It would be interesting to compare the used cell lines with early passages and the primary tumor tissue to examine this in more detail.
Taken together, these data suggest that HPV integration is not simply a random event, but rather has a preference for less protected and more accessible chromosomal regions such as transcribed tumor genes and fragile sites. It will be interesting to further explore (1) whether integration takes place in genes, which are highly expressed during carcinogenesis or (2) whether integration itself is rather random but may affect the expression of interrupted genes or (3) whether both may occur simultaneously. In this respect, Kraus Christiansen et al. (2015) recently reported that integration sites seem to coincide with DNA that is transcriptionally active in mucosal epithelium, as judged after relating data of integration sites to DNase hypersensitivity and H3K4me3 methylation. These results might point to integration being rather an early event in carcinogenesis than a late product of chromosomal instability, which is in agreement with data of Hopman et al. (2006) showing that integration already can occur in diploid CIN lesions.
4 Consequences of Viral Integration: Viral Gene Expression
In vitro studies have suggested that HPV integration events occur in cells that also contain non-integrated episomes resulting in repression of integrant-derived transcription of E6 and E7 by expression of the E2 transcriptional regulator from the episome (Bechtold et al. 2003; Pett et al. 2006; Groves and Coleman 2015). Only after episome clearance, for example by a host anti-virus response (Herdman et al. 2006), an upregulated expression of E6 and E7 oncoproteins from the integrated viral DNA might be detected, which leads to a selective growth advantage over cells harboring episomal DNA (Jeon and Lambert 1995). There is, however, discussion on the height of the E6 and E7 expression levels and how they are exactly regulated in HPV-positive lesions. The general view is that viral DNA often integrates in 1 or more copies into the host genomic DNA (see above). During this process, the viral episome is most often opened within the E2 open reading frame (preferential site of integration), frequently leading to deletion of E4 and E5 and part of E2 and L2 (zur Hausen 2002; Wentsenzen et al. 2004; Olthof et al. 2012). Olthof et al. and Parfenov et al. (2014) also detected disruption of the viral episome in the E1 gene, which also leads to E2 loss. The subsequent upregulation of E6 and E7 oncoproteins results in deregulation of cell signaling pathways, which, among others, leads to increased cellular proliferation and inhibition of apoptosis and finally to a transformed cell state (zur Hausen 2002; Ganguly and Parihar 2009; Moody and Laimins 2010; Pim and Banks 2010; Olthof et al. 2012) (Fig. 1b). Transformation is continuously dependent upon E6/E7 expression and can be reversed by the reintroduction of E2 (Adams et al. 2014) or by downregulation of E6/E7 using short-hairpin RNAs (Rampias et al. 2009). HPV breakpoints have also been mapped outside the E2 and E1 open reading frame (Akagi et al. 2014; Hu et al. 2015), most frequently in the L1 and L2 genes. In these cases, however, methylation of the E2-binding sites in the LCR promotor, preventing E2 to bind to the LCR promotor, might be responsible for de-repression of E6 and E7 expression (Reuschenbach et al. 2015). This might also be the case in tumors that harbor multiple copies of the HPV genome in stretches or concatenates in the human genome (Olthof et al. 2014; Groves and Coleman 2015). Another possibility might be that viral gene expression is influenced by nearby cellular regulatory sequences (Rusan et al. 2015).
In contrast to this view, a study in primary keratinocytes immortalized with HPV16 genomes has shown that disruption of the E2 gene sequence upon viral integration does not result in increased expression of the viral E6 and E7 oncogenes (Lace et al. 2011). In addition, a publication by Häfner et al. (2008) using APOT-PCR has shown no correlation between the integration state of the viral genome and the expression of the viral gene E6 in a collection of 55 HPV16-positive UCSCC samples. Recently, Olthof et al. (2014, 2015) have provided evidence that also in 7 HPV-positive HNSCC cell lines as well as in 75 primary OPSCC HPV physical status (extrachromosomal episomes or host DNA integrated) does not affect the levels of viral E2, E6 and E7 gene transcripts. Therefore, constitutive rather than a high-level expression of viral oncogene transcripts appears to be required in HPV-related OPSCC, enough to ensure the viral oncogenes to consistently deregulate cellular proteins and cell signaling pathways, including cell proliferation (pRb pathway), apoptosis and DNA damage response (p53 pathway) (Wiest et al. 2002; zur Hausen 2002; Hafkamp et al. 2009; Leemans et al. 2011; Pim and Banks 2010; Rieckmann et al. 2013; Arenz et al. 2014) (Fig. 1b).
5 Consequences of Viral Integration: Human Gene Expression
Besides its promotion of stable viral gene expression and subsequent deregulation of cell signaling pathways, HPV integration may also confer a selective growth advantage to the host cells through a direct effect on the host genome (i.e., by affecting the key cellular genes). Olthof et al. (2014) had mRNA expression profiling data of 6 OPSCC with proven HPV16 integration in gene sequences, including the known tumor-related genes FANCC, HDAC2, SYNPO2 and TRAF3. Viral integration, however, did not lead to significantly different expression of the interrupted gene in comparison with OPSCC having integration in another DNA sequence or showing solely viral episomes. This is in contrast to a study of Huebbers et al. (2013) showing that integration of low-risk HPV6 in the AKR1C3 gene resulted in loss of gene expression in a laryngeal carcinoma. In this case, however, the other gene copy was lost in the tumor as shown by array CGH analyses. In the 6 OPSCC studied by Olthof et al. (2014), no loss or amplification of the chromosomal regions containing the virally interrupted genes has been detected by array CGH, indicating that one or more expressed gene copies are still present in these tumors, which can mask a possible effect of the integration on gene expression. On the other hand, this might also point to the fact that viral integration is not per se meant to deregulate the interrupted gene in the cell, as also can be concluded by the finding of HPV16 integrated in intergenic sequences of 10 OPSCC in this study.
In UCSCC, however, Ojesina et al. (2014) found significantly elevated host gene expression levels at sites of integration compared with expression levels of the same genes in tumors without integration. This was associated in a number of cases with copy number gains, but not at all sites, indicating that expression may also be driven by alternative mechanisms, such as the viral promotor of the integrant, other regulatory sequences and proteins, or decreased E6/E7 expression (Rusan et al. 2015).
Figure 1c shows several mechanisms by which HPV integration may directly affect gene expression, previously presented by Rusan et al. (2015), i.e., (1) integration in a tumor suppressor gene resulting in loss of gene function, (2) integration adjacent to an oncogene leading to gene amplification and expression or enhanced expression from the viral promotor and (3) intra- or interchromosomal rearrangements followed by altered expression of genes in involved regions. Examples of (1) are described above and may involve additional loss of the chromosome without the HPV integrant (Huebbers et al. 2013) or amplification or loss of gene components leading to truncated proteins, as has been found for the double-strand break DNA repair pathway gene RAD51B (Khoury et al. 2013; Ojesina et al. 2014; Parfenov et al. 2014). HPV integration upstream near or within the NR4A2 or MYC oncogenes in UCSCC and OPSCC are examples of (2) (Ferber et al. 2003; Wentsenzen et al. 2004; Ojesina et al. 2014; Parfenov et al. 2014), and examples of HPV insertion associated with chromosomal rearrangements, gene amplification and increased expression have been described by Akagi et al. (2014), Parfenov et al. (2014) and Olthof et al. (2015) involving the TP63 gene, a transcription factor with a role in epithelial development and highly expressed in squamous cell carcinomas (SCC).
In summary, recent as well as older literature has provided evidence that at least in a part of UCSCC and OPSCC HPV integration has a direct effect on the host genome and human gene expression, further underscored by recurrent integration events in specific genes. However, more studies are needed to fully explore the molecular mechanisms underlying human as well as viral gene expression as a result of HPV integration in anogenital and head and neck cancers.
6 HPV Integration in Relation to Viral Load, Methylated Genes and Outcome
A number of studies have examined other parameters in relation to HPV integration, although different methods have been used to determine the viral physical status. Olthof et al. (2014) examined whether tumors with episomal virus have a higher viral load than those with integration as determined by APOT and/or DIPS-PCR. For this purpose qPCR was performed on 73 OPSCC samples. Viral load ranged from 3.4 × 10−6 up to 97 HPV DNA copies per cell. When comparing the average viral load in cases with or without integration, no significant differences were seen (7 vs. 8.5 HPV DNA copies/cell). Furthermore, no correlation was found between the mean log2 expression levels of the viral genes E2, E6 or E7 and the viral load. This was also the case in 7 HPV16-positive HNSCC cell lines containing 2–7 integration sites, in which the viral load ranged from 1-739 HPV DNA copies/referencee gene (Beta-globin) copy (Olthof et al. 2015).
In two studies, methylation of human genes as well as E2-binding sites in the HPV LCR DNA, respectively, were examined and compared with the HPV integration status of head and neck cancers. In the first study, Parfenov et al. (2014) showed that DNA methylation profiles are distinct for HPV-positive tumors with integration than for those without integration. Differentially methylated genes included the tumor suppressors BARX2 and IRX4, and the oncogenes SIM2 and CTSE. The mechanism by which integration alters the methylation profile, however, remains to be elucidated (Rusan et al. 2015). In the second study, Reuschenbach et al. (2015) detected differential methylation levels in the HPV16 (LCR) E2-binding sites E2BS3 and E2BS4 depending on the viral DNA physical status, i.e., (1) complete methylation (>80 %) associated with the presence of integrated HPV genomes with an intact E2 gene; (2) intermediate methylation levels (20–80 %) with predominantly episomal HPV genomes with intact E2; and (3) no methylation (<20 %) with a disrupted E2 gene. Patients with high methylation levels tended to have a worse 5-year overall survival compared with patients with intermediate methylation (hazard ratio: 3.23). The authors therefore concluded that further studies are warranted to determine whether the E2BS methylation status may represent a prognostic marker.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree