Keywords
Oncolytic virus, molecular imaging, bioluminescence imaging, fluorescence imaging, PET, SPECT
Preclinical Imaging of Viral Gene Expression
One important reason for the ongoing and increasing success of oncolytic viruses in the treatment of cancer is their ability to target and destroy the tumor by multiple mechanisms . This includes (1) the direct oncolytic effects of tumor cell destruction as a result of selective viral replication in the cancer and (2) viral-mediated induction of an immune response against the tumor, including transient overcoming of localized immune suppression within the tumor and induction of an antitumor adaptive immune responses through in situ vaccination effects . In addition, recently, the phenomenon of viral-mediated vascular collapse locally within the tumor has been described mediated by viral-induced thrombosis and direct targeting of endothelial cells within the tumor . All of these and additional mechanisms of tumor killing can be further enhanced through transgene expression from the viral vectors. However, the capacity to simultaneously explore the effects of these multiple mechanisms of action in preclinical studies requires animal models. Furthermore, most of the common reasons for failure of oncolytic viruses will only be seen in the context of an intact organism, including induction of too rapid an immune response targeting the virus or failure of delivery in the face of preexisting antiviral immunity; an inability of the vector to successfully escape circulation and enter the tumor; reduced capacity to spread efficiently within the tumor due to barriers produced by stromal cells and tumor matrix; or inefficient selectivity, leading to toxicity in essential organs, which is likely to be missed in cell culture studies, in which it is impossible to screen every nontumor cell type.
However, animal testing studies are expensive and laborious, and traditionally there has been only a limited capacity to follow events in a living animal beyond external measurement of the size of artificial subcutaneous tumors or observation of surrogate measures of toxicity and pathogenicity such as weight loss. Alternatively, the use of endpoint studies requires sacrifice of cohorts of multiple animals at many time points and also postmortem analyses requiring extensive processing of multiple tissues. Even then, it is possible that key times and organs may be missed. It is therefore clear that approaches that permit a more rapid and global examination of an oncolytic virus in vivo and allow the study of its interactions with selected components of the host organism or tumor can benefit preclinical development of oncolytic virotherapy, especially noninvasive or minimally invasive approaches that allow repeated imaging of the same animals during the course of a treatment. Noninvasive, whole animal molecular imaging of reporter transgene expression is a powerful tool that can provide precise and unique spatiotemporal measurements of viral replication in vivo .
One advantage of many oncolytic viral therapies is the ability to express transgenes from the viral backbone. This has been tested extensively with therapeutic genes used to arm the virus and enhance their antitumor effects. However, recently it has become common to also express reporter genes whose expression can be imaged from within whole animals or from cancer patients in the clinic.
Typically, the reporter genes and imaging modalities that have been used most extensively in the preclinical development of oncolytic vectors differ from those used in the clinic. The more clinically relevant molecular imaging modalities used in cancer diagnosis , such as radiological imaging (including positron emission tomography (PET) and single-photon emission computed tomography (SPECT)) and magnetic resonance imaging (MRI), tend to require more expensive equipment and specialist trained operators, which means that their preclinical use is usually restricted to development of imaging approaches for direct clinical translation. Instead, the majority of noninvasive, whole animal molecular imaging incorporated into preclinical studies utilizes optical modalities . Optical imaging techniques (bioluminescence and fluorescence imaging) do not currently have direct clinical counterparts, primarily due to limitations in depth penetration of the imaging; however, they are sensitive and tend to be relatively inexpensive, rapid, and straightforward to run, making them ideal for proof-of-concept studies. They are therefore discussed in detail.
Bioluminescence Imaging
Bioluminescence imaging (BLI) relies on the detection of visible photons of light released at specific wavelengths through energy-dependent reactions catalyzed by luciferase enzymes . Several families of luciferase enzymes are found naturally , and many of these have been adapted for use in BLI through mammalian codon optimization and removal of phantom transcription initiation sites. These include insect luciferases (e.g., from the firefly or click beetle ), marine luciferase (e.g., Renilla luciferase from the sea pansy), and bacterial luciferases (e.g., luciferase from the symbiotic bacteria Vibrio fischeri ). These enzymatic reactions require the gene to be expressed in the cell of interest and also exogenous delivery of a substrate (luciferin for the insect luciferases or coelenterazine for marine luciferases), whereas the insect luciferases also require ATP, magnesium, and oxygen . This means that the insect luciferase enzymes, unlike fluorescent proteins, will typically only function within a living cell. Also, because the luciferase enzymes are relatively unstable (with a half-life on the order of several hours), bioluminescence measurement provides a good correlation with active gene expression, unlike many fluorescent proteins that may be very stable and remain present in a cell for days after active expression has ceased. As a result, viral expression of luciferase provides a robust correlation to active replication .
The bioluminescence emitted can be detected and amplified using specialized cameras with highly sensitive detection systems , revealing the sites and levels of luciferase expression and activity from within a living animal. However, this approach is limited by the properties of light passing through tissue, which leads to diffraction and absorption . As a result, imaging is only possible up to a depth of approximately 1 or 2 cm. This limits BLI applications for anything other than surface imaging to rodent animal models . In addition, the diffraction of light limits the resolution achievable to slightly less than 1 mm. Because light in the far-red and near-infrared range (between 600 and 900 nm) has the greatest ability to pass through tissues , it is important that luciferases produce light spectra in this window. By virtue of this wavelength of light produced and the solubility, stability, and membrane permeability of its substrate , the luc luciferase from firefly species ( Photinus ) has become the most commonly used luciferase for in vivo imaging. Although the separate substrate used by Renilla luciferase permits the imaging of two luciferases in a single animal , this approach is rarely used due to the inappropriate wavelength of light produced by the Renilla enzyme and the instability of its substrate . Because of the rapid imaging of multiple animals, ease of use of the imaging instrumentation, and inexpensive nature of bioluminescence imaging, it has become a powerful research tool for the preclinical development of many novel therapies, especially for cancer.
The applications of BLI in biomedicine are extensive. It has been used for tracking bacterial pathogens , to study gene expression patterns , to monitor tumor cell growth and regression , to determine the location and proliferation of stem cells , and to track gene expression patterns . In the context of oncolytic viruses, bioluminescence has been used primarily to monitor viral replication in vivo and biodistribution in preclinical models ( Figure 31.1 ). The luciferase gene has been cloned and used for tracking the replication of oncolytic parvoviruses , adenoviruses , HSV-1 , vaccinia virus , measles virus , and vesicular stomatitis virus (VSV) . Furthermore, the infection of carrier cells with oncolytic viruses expressing luciferase can be used to evaluate the biodistribution of those cells. This strategy was used to evaluate the biodistribution of carrier CIK (cytokine-induced killer) cells loaded with vaccinia virus expressing luciferase or T cells loaded with measles virus . As an alternative to following biodistribution, levels, and persistence of viral gene expression in whole animal models, it is also common to label the tumor cells with the luc gene and so quantify the kinetics and levels of response to treatment and relapse of the tumor . This is especially useful for orthotopic or peritoneal implantation of tumors, and several spontaneous tumor models have also been adapted to allow for luciferase expression exclusively from within the tumor cells. However, one limitation of BLI is the restriction to imaging of a single reporter at any one time, which means that simultaneous imaging of tumor and virus within a single animal, although feasible, is usually impractical.
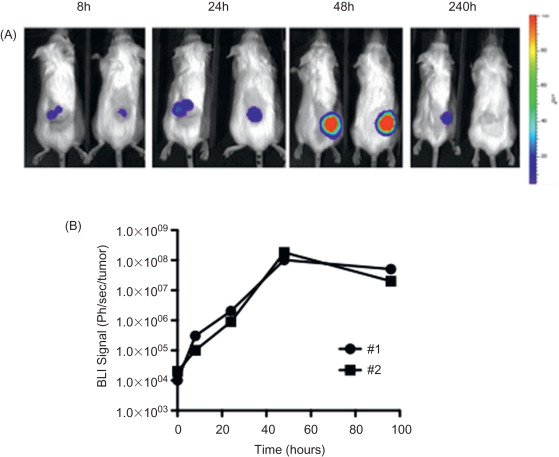
In addition to its use for understanding the activity and action of novel vectors in preclinical models, BLI has also been used to define the potential usefulness of imaging of viral gene expression in a clinical setting (even though alternative imaging modalities would subsequently need to be incorporated). For example, imaging of luciferase-expressing oncolytic viruses has been used to predict the therapeutic outcome of the oncolytic therapy. In preclinical models, the predictive value of an early time point imaging of adenoviral replication in tumors has been demonstrated . The in vivo luciferase bioluminescence measured 6 days after viral administration significantly correlated with the antitumor effect observed on day 36. Alternatively, it has also been proposed that imaging of oncolytic viral gene expression might be used to reveal the locations of primary tumors and metastases. It was shown that an intravenously delivered oncolytic vaccinia virus expressing luciferase replicated in the tumor tissue and permitted the delineation of the location of tumors and metastases . However, it remains to be seen if delivery would be sufficiently reliable to locate all metastases using this approach in a clinical setting. Despite the fact that luciferase expression and BLI cannot currently be used in humans, these studies provide proof-of-concept that oncolytic viruses expressing clinical-friendly imaging systems may also improve the outcome of clinical protocols.
Fluorescence Imaging
Fluorescent proteins have also been extensively used as reporter genes expressed from oncolytic viruses, especially for the purpose of tracking the replication of the virus in vitro . Fluorescent proteins absorb a photon of light of a set wavelength (excitation light) before releasing the photon (emission light) with reduced energy at an increased wavelength (through a Stokes shift), thus allowing accurate determination of the localization and concentration of the fluorophore through quantification of emission light released after excitation . Because no substrate and no additional energy source are required, fluorescence imaging is simple and adaptable, and because of the capability to distinguish fluorophores based on their excitation/emission spectra, the imaging of multiple fluorophores is possible. The expression of fluorescent proteins therefore offers another possibility to image viral replication rapidly and inexpensively.
The fluorescent protein most commonly cloned into oncolytic viruses is green fluorescent protein (GFP), a 27-kDa protein from the jellyfish Aequorea victoria that fluoresces green upon illumination with ultraviolet light . This wild-type protein and its enhanced-fluorescent form have been cloned into the majority of oncolytic viruses in which genetic manipulation of the viral genome is possible, including Newcastle disease virus , VSV , HSV-1 , measles , adenovirus , and vaccinia virus .
However, despite the power of imaging of GFP expression from oncolytic viruses in cell culture, the use of GFP for monitoring virus replication within organs in living animals is limited due to the very low tissue penetrance of both excitation and emission wavelengths of light required for GFP imaging; thus, imaging depth penetration is often limited to 1 or 2 mm. Despite this limitation, groups have reported successful imaging of oncolytic viruses expressing GFP in mice, such as a GFP-expressing Newcastle disease virus, demonstrating the possibility of using this protein for in vivo imaging, even though accurate quantification is likely to remain an issue . Alternatively, approaches such as fiber-optic monitoring coupled with confocal microscopy to allow direct, rapid, and sensitive visualization of fluorescent signals in organs such as the brain have been used, but the application of intravital microscopy is typically highly invasive .
To allow the routine, sensitive, and quantifiable application of fluorescent imaging noninvasively to small animals, novel strategies have been developed. These tend to be classified into two categories, with improvements in both the imaging apparatus and the fluorescent proteins. For example, complex and elegant genetic engineering of the GFP gene has resulted in red-shifting of the excitation and emission wavelengths, resulting in fluorescent proteins that fluoresce in the far-red and near-infrared spectrum . These maximize tissue penetrance and minimize the level of autofluorescence. Proteins such as the far-red fluorescent proteins TurboFP635 or iRFP have the potential to be expressed from oncolytic viruses or tumor cells and imaged using advanced imaging methods in whole animals in three dimensions, with the additional advantage that different excitation lasers and filter sets will allow the imaging of multiple fluorescent proteins or fluorescent probes in a single animal, thus allowing the simultaneous interrogation of multiple parameters.
Advances in imaging technology, such as fluorescence molecular tomography (FMT) , have also allowed for recent advances in the capabilities of applying fluorescence imaging to whole animals. In FMT, the subject is exposed to continuous-wave or pulsed light, and the emitted light is captured by detectors in an imaging chamber arranged in a spatially defined order . A tomographic image is reconstituted after the data are mathematically processed, and quantitative, three-dimensional molecular information can be extracted.
In addition to monitoring virus replication, oncolytic viruses expressing fluorescent proteins have been tested preclinically for a variety of putative clinical applications. As with luciferase-expressing viruses, oncolytic viruses expressing fluorescent proteins may also be applied to predict therapeutic outcome. One method becoming routinely used is the infection of explant tissues from patients ex vivo prior to therapy in order to ascertain the likelihood of successful oncolytic virotherapy or to define cancers most susceptible to a vector. Demonstrating functionality of the oncolytic agent via fluorescence imaging within ex vivo tissues may be a useful bridge between preclinical studies and clinical trials. Alternatively, GFP-expressing viruses have also been proposed for precise surgical navigation due to its ability to infect and express GFP selectively in cancer cells. A GFP-expressing adenovirus has been used for fluorescence-guided surgery in a model of intraperitoneal disseminated colon cancer and in a model of pleural disseminated lung cancer . Five days after virus intraperitoneal or intrapleural administration, a fluorescence-guided laparotomy permitted the resection of disseminated cancer nodules, which would otherwise be undetectable. An HSV-1-expressing GFP was also used for this purpose. In a model of peritoneal carcinomatosis, the administration of the oncolytic virus 48 hr prior to surgery allowed the detection of residual disease in 8 of 13 mice that experienced presumably complete cytoreduction by surgeons . Residual disease was identified in sites corresponding to patterns of recurrence in a published human series, highlighting the potential of combining surgery with virally directed fluorescence imaging. Moreover, this technique has also been used for the detection of lymph node metastases. In a model of metastatic melanoma, the administration of an engineered vaccinia virus expressing GFP into the primary tumor resulted in viral transmission to lymph nodes, infection of lymphatic metastases, and transgene expression that was reliably and easily detected . The real-time fluorescence detection of nodal disease was 80% sensitive and 100% specific for identification of sentinel node disease. In a model of metastatic breast cancer, the injection of a GFP-expressing HSV-1 directly into primary breast tumors also resulted in viral transit to axillary lymph nodes, infection of lymphatic metastases, and GFP expression . This technique permitted the real-time intraoperative identification of micrometastases otherwise undetectable due to the impracticality of performing extensive histologic analyses on a large number of resected nodes.
Clinical Imaging of Viral Gene Expression
Because the optical imaging modalities commonly incorporated in preclinical studies do not have clinical equivalents, the translation of imaging of viral replication into the clinical setting has required switching of reporter genes, predominantly to those suitable for nuclear medicine-based imaging (PET and SPECT). As most other clinical imaging modalities are primarily structural (computed tomography (CT), X-ray, or ultrasound)—that is, they do not readily support the molecular imaging required to visualize gene expression. However, MRI does have the potential to visualize gene expression (e.g., through expression of ferritin genes ), but to date this approach has not been applied to oncolytic viruses.
The primary limitation of nuclear medicine approaches is that because of the reduced sensitivity relative to optical imaging, choice of reporter and associated radioisotope is critical. As a result, extensive preclinical testing in animal models is still required. To date, the majority of PET- and SPECT-based imaging of oncolytic viruses has occurred in preclinical models, but sufficient clinical data to encourage further development have been reported.
Nuclear Medicine-Based Imaging
Nuclear medicine-based imaging is reliant on the ability of scanners to detect and localize gamma ray emission from the decay of a radiotracer. The scanners are sensitive, provide good spatial resolution, and allow for imaging with far greater tissue penetration than optical approaches. However, because these approaches are typically less sensitive than optical imaging and require the use of more expensive cameras and radioactivity, they are not routinely used for preclinical virotherapy development . The gamma camera and PET and SPECT scanners are the principal types of scanners used in nuclear medicine. Because the images produced provide no structural references, this imaging is typically performed in conjunction with a second modality, such as X-ray or CT, to provide detail of the localization of signal. When applied to oncolytic viruses expressing a reporter transgene, viral replication can be monitored as a result of the alteration of the biodistribution of a tracer molecule at the sites of expression of the reporter gene, leading to a local concentration of the tracer at the site of viral replication.
Three types of reporter gene families have been cloned into oncolytic viruses. In the first family, the accumulation of the radiolabeled substrate is based on enzymatic activities that trap the tracer inside target cells when phosphorylated or processed by the enzyme. For example, when working with HSV-1, the endogenous thymidine kinase can be used for this purpose. FIAU and [ 18 F]FHBG have been used previously as the substrate for HSV-TK to successfully image viral replication in animal models using a PET scanner . Furthermore, a Sindbis virus expressing HSV-TK was also successfully imaged using 18 F-FIAU as a substrate with a PET scanner . However, in an attempt to monitor the replication of an oncolytic HSV-1 in glioma patients, no evidence of viral replication was found using a 123 I-FIAU brain SPECT scanner . This unsuccessful result was probably due to limited expression of endogenous HSV-TK and limited sensitivity of 123 I and SPECT imaging, indicating the need for higher reporter gene expression, more potent reporter genes, and/or greater sensitivity in radioisotope detection for the clinical setting.
The second family of reporter genes involves membrane receptors that bind and trap a radiolabeled substrate. The human somatostatin receptor SSTR2 was successfully used to monitor the replication of an oncolytic vaccinia virus in mice using 111 In-pentetreotide as ligand and a gamma camera for the imaging . Furthermore, the dopamine DR2 receptor with 123 I-IBF or 11 C-raclopride is also suitable for application to oncolytic viruses .
Finally, the third and most extensively examined family is that of membrane proteins that mediate ionic transport. The human norepinephrine transporter (hNES) has been used in the context of an oncolytic vaccinia virus to monitor its replication in mice , but the most broadly used ion transporter used for imaging is the human sodium iodide symporter (hNIS). This can be imaged using [ 123, 124, 131 I]NaI, [ 99m Tc]pertechnetate, or [ 186, 188 Re]perrhenate, among others, and using scintigraphy, PET, or SPECT scanners . To monitor viral replication in preclinical models, it has been cloned into measles virus , VSV , adenovirus , and vaccinia virus . In the clinical setting, the feasibility of using hNIS-mediated imaging was demonstrated in a phase I clinical trial treating prostate cancer patients with an oncolytic adenovirus coding for the hNIS gene . In this study, viral replication was detected in seven of nine patients when 1×10 12 virus particles were injected. However, the replication of a second oncolytic adenovirus expressing the hNIS gene failed in a clinical setting . In this study, a cervical metastatic carcinoma patient was treated with 3×10 11 virus particles and a SPECT-CT scanner was used for the imaging. The difference in the success of the imaging is likely explained by the dose administrated, the virus design, and especially the relative sensitivity of PET and SPECT scanners.
Alternative applications for the combination of nuclear medicine-based imaging with oncolytic viruses have also been described. As with oncolytic viruses expressing fluorescent proteins, the endogenous thymidine kinase coded by an oncolytic herpes virus has been used for the imaging and detection of lymph node micrometastases. In a preclinical model of metastatic melanoma, eight of eight tumor-positive nodes were successfully identified after an intratumoral injection of the oncolytic agent . No overlap between radioactivity levels in tumor-positive and tumor-negative lymph nodes using [ 18 F]FEAU PET was described. Furthermore, genes such as hNIS can be used for radiotherapy. In addition to imaging, the radiation physical crossfire effect permits the destruction of noninfected tumor by radiation emitted from neighboring cells transduced by recombinant oncolytic viruses . This effect is of relevance because it allows for the destruction of intratumoral physical barriers such as stromal cells that can otherwise handicap the dispersion of the virus throughout the tumor. Notably, results from xenograft tumors treated with an oncolytic measles virus expressing NIS showed complete regression of the tumor after 131 I therapy .
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree