Fig. 12.1
One major issue with learning from images is the large dimensionality of the raw data. Even if a smaller region of interest (ROI) is used, each ROI image typically has a dimensionality of 10,000. In addition, high-dimensional data also requires increased computational cost, which creates problems for real-time applications such as respiratory gating. Thus, it is crucial to apply dimensionality reduction techniques to images before any learning procedure is performed.
Cui et al. [2] presented the first study using machine learning approaches for respiratory gating. In their approach, the fluoroscopic images acquired during patient setup are first transformed into a lower dimensional space using principal component analysis (PCA) for training purposes. These samples with class label are used to train a classifier based on support vector machine (SVM). After the optimal classifier is determined, new images are acquired during treatment delivery, which are projected to the same PCA feature space, and passed to the SVM classifier obtained in the training session. The output of the classifier is the predicted label of the new image, which determines whether to turn the beam on or off at any given time. When tested on five sequences of fluoroscopic images from five lung cancer patients, the SVM classifier was found to be slightly more accurate on average (1–3 %) than the template matching method, and the average duty cycle is 4–6 % longer.
In a follow-up study, Lin et al. [15] performed more comprehensive evaluations of different combinations of dimensionality reduction and classification techniques. They investigated four nonlinear dimensionality reduction techniques, including locally linear embedding (LLE), local tangent space alignment (LTSA), Laplacian eigenmap (LAP), and diffusion maps (DMAP). For classification, a three-layer artificial neural network (ANN) was used in addition to SVM. Performance was evaluated on ten fluoroscopic image sequences of nine lung cancer patients. It was found that among all combinations of dimensionality reduction techniques and classification methods, PCA combined with either ANN or SVM achieved a better performance than the other nonlinear manifold learning methods. ANN when combined with PCA achieved a better performance than SVM, with 96 % classification accuracy and 90 % recall rate, although the target coverage is similar at 98 % for the two classification methods. Furthermore, the running time for both ANN and SVM with PCA is around 6.7 ms on a Dual Core CPU, within tolerance for real-time applications. Overall, ANN combined with PCA was found to be a better candidate than other combinations for real-time gated radiotherapy.
In the above previous works, PCA was used as a dimensionality reduction technique to preprocess the data. In [12] the generalized linear discriminant analysis (GLDA) was applied to the respiratory gating problem. The fundamental difference from conventional dimensionality reduction techniques is that GLDA explicitly takes into account the label information available in the training set and therefore is efficient for discrimination among classes. On average, GLDA was demonstrated to perform similarly with PCA trained with SVM at high nominal duty cycles and outperform PCA in terms of classification accuracy (CA) and target coverage (TC) at lower nominal duty cycle (20 %). A major advantage of GLDA is its robustness, while CA and TC using PCA can be reduced by up to 10 % depending on the data dimensionality. With only 1-dimensional feature vectors, GLDA is much more computationally efficient than PCA. Therefore, GLDA is an effective and efficient method for respiratory gating with markerless fluoroscopic images.
12.3 Real-Time Tumor Tracking Based on Fluoroscopic Images
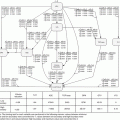
Since the output of a real-time tumor tracking system is a continuous variable, it can be formulated as a regression problem from a machine learning perspective. Lin et al. [14] proposed to use learning algorithm for tumor tracking in fluoroscopic images, based on the observation that the motion of some anatomic features in the images may be well correlated to the tumor motion (Fig. 12.2). The correlation between the tumor position and the motion pattern of surrogates can be captured by regression analysis techniques. The proposed algorithm consists of four main steps: [1] selecting surrogate regions of interest (ROIs), [2] extracting spatiotemporal patterns from the surrogate ROIs using PCA, [3] establishing regression between the tumor position and the spatiotemporal patterns, and [4] predicting the tumor location using the established regression model. In a clinical setting, the first three steps would be performed using training image data before the treatment, while the final step would be performed in real time using the image data acquired during treatment delivery.
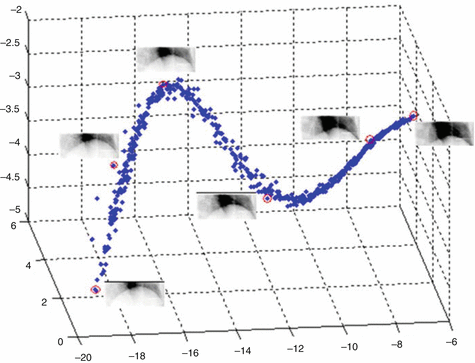

Fig. 12.2
3D embedding of the diaphragm ROI images using PCA. Representative images are shown next to circled points at different location in the 3D PCA space, representing different positions of the diaphragm (Reprint from Lin et al. [14])
They evaluated several regression techniques for tracking purposes, including linear regression, second-order polynomial regression, ANN, and SVM. The experimental results based on fluoroscopic sequences of 10 lung cancer patients demonstrate a mean tracking error of 1.1 mm and a maximum error at a 95 % confidence level of 2.3 mm for the proposed tracking algorithm. The results suggest that the machine learning approaches are promising for real-time tumor tracking. However, these methods have to be fully validated before their clinical use. In particular, PCA is sensitive to the tumor size and position, so if the tumor changes size or relative position with respect to the chosen surrogates, the regression model needs to be re-evaluated. This suggests that a separate training data set may be required for each treatment fraction for the learning technique to work well.
Li and Sharp [13] proposed a fluoroscopic fiducial tracking method that exploits the spatial relationship among the multiple implanted fiducials. The spatial relationships between multiple implanted markers are modeled as Gaussian distributions of their pairwise distances over time. The means and standard deviations of these distances are learned from training sequences, and pairwise distances that deviate from these learned distributions are assigned a low spatial matching score. The spatial constraints are incorporated in two different algorithms: a stochastic tracking method and a detection-based method. In the stochastic method, hypotheses of the “true” fiducial position are sampled from a pre-trained respiration motion model. Each hypothesis is assigned an importance value based on image matching score and spatial matching score. Learning the parameters of the motion model is needed in addition to learning the distribution parameters of the pairwise distances in the proposed stochastic tracking approach. In the detection-based method, a set of possible marker locations are identified by using a template matching-based fiducial detector. The best location is obtained by optimizing the image matching score and spatial matching score through non-serial dynamic programming. The proposed method was evaluated using a retrospective study of 16 fluoroscopic videos of liver cancer patients with implanted fiducials. On the patient data sets, the detection-based method gave the smallest error (0.39 ± 0.19 mm). The stochastic method performed well (0.58 ± 0.39 mm) when the patient breathed consistently; the average error increased to 1.55 mm when the patient breathed differently across sessions.
12.4 Real-Time Tumor Tracking via Volumetric Imaging Based on a Single X-Ray Image
Li et al. [9] have recently made a breakthrough in reconstructing volumetric images and localizing lung tumors in real time using a single x-ray projection image. The method is based on an accurate patient-specific lung motion model and uses the CT images acquired during simulation as the reference anatomy. For lung cancer patients, a respiration-correlated 4DCT is typically acquired for treatment simulation purposes. Deformable image registration (DIR) is performed between a reference CT image and all other CT images, and a set of displacement vector fields (DVFs) will be obtained, which basically tells how each voxel/point in the lung moves, or its 3D motion trajectory. Given the dense DVFs, a patient-specific lung motion model is built based on PCA. The PCA motion model is accurate, efficient, and flexible and imposes implicit regularization on its representation of the lung motion [11]. As a result, a few scalar variables (i.e., PCA coefficients) are sufficient in order to accurately derive the dynamic lung motion for a given patient. Therefore, limited information, e.g., a single x-ray projection, can be used to reconstruct the volumetric image of the patient anatomy, in which the PCA coefficients are optimized such that the projection of the reconstructed volumetric image corresponding to the new DVF matches with the measured x-ray projection. Once the optimal DVF has been found, the 3D tumor location relative to the reference position defined in 4DCT can be determined. The algorithm was implemented on graphic processing unit (GPU). The average computation time for image reconstruction and 3D tumor localization from an x-ray projection ranges between 0.2 and 0.3 s on the C1060 GPU card.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree