Technique
Resolution
Sensitivity
Purification
Loading capacity
Protein/peptide identification
Limitations
SELDI-TOF
Low
Low
Not required
Low
No
No peptide identification
nLC-MS/MS
Medium
Low
Required
Low
Yes
Limited loading capacity
2DE-MS
Low
Medium
Required
Low
Yes
Reproducibility
Ultrafiltration – MS
Medium
Low
Not required
Medium
Yes
Interference by high mw proteins
1D SDS-PAGE–LC-MS/MS
High
High
Not required
High
Yes
Low throughput
ICAT-MS
High
Medium
Required
Low
Yes
Radioactive
A surge of interest in defining molecular biomarkers of health and disease from blood plasma has emerged with the conclusion of the pilot phase of Plasma Proteome Project (PPP) by Human Proteome Organization (HUPO) (Omenn et al. 2005). Alongside multiple other allied projects to profile the human plasma proteome including PeptideAtlas, GPMO have contributed their share (Omenn et al. 2005; Liu et al. 2007; Saha et al. 2008; Deutsch et al. 2005). Most commonly used approach in plasma proteomics is to fractionate the intact protein mixture and study each fraction separately to maximize the depth of coverage.
In sections to follow, we will confine our discussion to the albuminome and sub-proteomes it directly influences; in particular the lower molecular weight proteins and low-abundance proteins identified by single peptide hits while considering the current challenges and future directions in its characterization.
9.2 Sub-proteomes in Blood Plasma
Plasma represents a super proteome where all other sub-proteomes merge. This super proteome of body is linked with all other proteomes of body organs such as liver proteome, brain proteome, and CSF proteome, which are connected by means of tissue fluid and lymph. Consequently the natural sub-proteomes of plasma such as low molecular weight proteome, albuminome, plasma metabolome, and disease-specific degradome are under keen observation as they may represent the direct status of pathophysiology of a disease (Villanueva et al. 2006). While the ease of availability of plasma has always been an important factor for the keen research interest in this clinical sample, majority of advancements in this field have been made during the past decade. Most of this can be attributed to simultaneous development and improvement in the fractionation strategies and proteomic technologies. Depending upon the desired end points, a number of strategies have been explored and have reported successful application in the area of plasma/serum proteomics.
9.3 Albuminome: Trash or Biomarker’s Diamond Mine
Albumin is the oldest known protein in human history with almost 230 years history. Albumin (66.5 kDa) is non-glycosylated and most abundant plasma protein with six strong binding sites – I and II for small organic molecules, III and IV for long chain fatty acids, V for cys, and VI for the square-planar N-terminal Cu2+ & Ni2+ sites (Christodoulou et al. 1995). For decades, the research in field of blood plasma and serum was focused on removal of albumin to visualize the low-abundance proteins. This trashed fraction however was actually a treasure in form of albumin that contained a large number of molecular species reflective of pathophysiology of an individual. It is well known that albumin binds proteins and peptides and peptide hormones such as serum amyloid A, interferons, glucagons, bradykinin, insulin, and protein G (Sjöbring and Kastern 1991; Steel et al. 2003). However, not until recently has a comprehensive proteomic investigation into serum albumin and its bound proteins/peptides been conducted.
9.3.1 Lower Molecular Weight Proteome
Lower molecular weight (LMW) fraction of plasma proteins includes proteins with less than 30 kDa molecular weight. This is of particular interest in case of cancers as tumor microenvironment has been shown to differentially express the exoproteases leading to cancer-specific proteolytic activity and altered post-translational modifications resulting in production of LMW proteins and peptides from larger molecules (Villanueva et al. 2006). The LMW fraction of plasma largely remains uncharacterized until recently due to the renal clearance limit of 30 kDa and its variability and more importantly due to the pre-fractionation procedures used (Maack et al. 1979; Tirumalai et al. 2003; Adkins et al. 2002).
Partial denaturation of plasma leads to enrichment of LMW fraction and results have supported the identification of proteomic patterns observed in case of breast and ovarian cancers (Merrell et al. 2004; Huang et al. 2005). A number of the potential biomarkers for detection of cancer have been reported in the recent past, majority of these are represented by the LMW proteins such as Fibrinopeptide A for hepatocellular carcinoma (Orvisky et al. 2006), serum amyloid A in renal cancer (Tolson et al. 2004), and kallikreins in prostate cancer. Apart from the intact LMW proteins, a number of proteolytic products of higher MW proteins also contribute to the LMW proteome of plasma. This includes derivatives of complement proteins, IgG, enzymes, transport proteins, and tissue protein derivatives (Fig. 9.1).
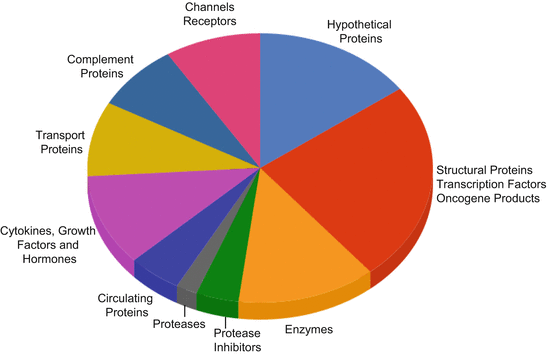

Fig. 9.1
Distribution of proteins in the LMW plasma proteome. Pie chart representing the widespread distribution of LMW proteins in circulatory proteome from structural proteins to proteases and oncogene products (Redrawn from (Tirumalai et al. 2003))
Technical issues, particularly reproducibility, have hampered the large-scale analysis of LMW proteome. In the past, removal of high-abundance proteins, centrifugal filtration, and IEF have been employed with adequate success but were not found to be reproducible. Not until recently, albumin was explored as a source for the detection of these LMW proteins due to its properties of carrier protein.
9.3.2 Kinetics of Albumin Binding to LMW Proteins
The renal clearance limit of globular proteins in humans is 30 kDa and theoretically any protein/peptide below this limit is rapidly cleared by glomerular filtration in kidneys. The concentration of a biomarker is function of the ratio between biomarker production rate from the tissue and rate of clearance of the carrier protein which in this case is albumin. Consequently, albumin binding amplifies the total biomarker concentration levels measured in serum or plasma. Amplification occurs because the carrier protein acts as a reservoir to accumulate the biomarker over time, as the tissue is continuously producing the biomarker. Thus, a biomarker produced by a small volume of tissue such as the ovary (Petricoin et al. 2002a, b), prostate (Petricoin et al. 2002c), or breast (Li et al. 2002), at a low concentration (e.g., one femtomole per day), can accumulate to a concentration of one picomole in plasma because it binds with albumin that has long half-life of 6 days. Figure 9.2 shows the mechanism of amplification of LMW proteins by binding to albumin which functions as reservoir for the accumulation and amplification of low mass biomarkers produced during cancer progression.
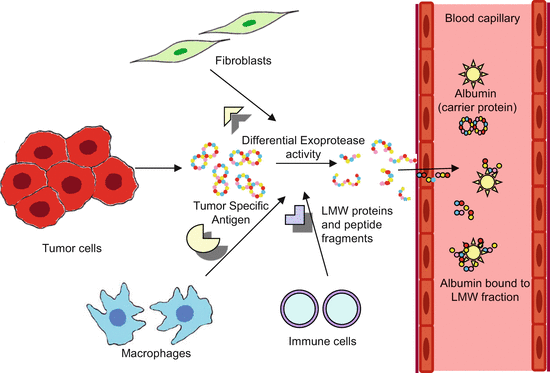

Fig. 9.2
Albumin binding and amplification of LMW proteins. As a result of tumor progression and tissue-specific exoprotease activity in the tumor microenvironment, multiple protein isoforms of biomarker proteins are formed. These potential biomarkers traverse through the endothelial lining of the newly formed tumor vasculature and enter the blood circulation and bind to the carrier proteins like albumin, protected from renal clearance and are amplified
Kinetics of albumin association to the lower molecular weight biomarkers have revealed that binding to albumin may affect the protein’s availability, detection, quantization, and overall utility as a biomarker (Mehta et al. 2003). Additionally, the presence of unbound and bound states of proteins in plasma can affect both the clearance of free-state proteins/peptides and also their detection by current proteomic approaches. Further, pattern of albumin bound proteins is distinct from those bound to other proteins (Mehta et al. 2003). Novel protein hits continue even over five iterations indicative of vast repertoire of albumin-associated proteins. It is therefore imperative that examination of the LMW species bound to albumin may contain important diagnostic information for neoplastic transformation.
9.3.3 Albuminome
Albumin and its bound proteins and peptides are referred to as albuminome that is a naturally occurring sub-proteome in blood. A robust, optimized, and reproducible chemical method for the isolation of albumin from serum based on classical Cohn fractionation has recently been used (Fu et al. 2005). Chemical fractionation eliminated the problem of protein carryover in subsequent experiments and can be extended to any volume of plasma and was compatible with both 2DE and LC-MS/MS. From the reported presence of 120 nonredundant proteins in the albumin-enriched fraction – albuminome – majority (75 %) were extracellular proteins, whereas the minority (25 %) were cellular proteins (Fu et al. 2005). This latter category contains transport proteins, enzymes, and protease inhibitors. Although many of the proteins present in the albumin-enriched fraction are high-abundance proteins, such as α1-antitrypsin and transferrin, lower-abundance proteins such as fibrinogen and angiotensinogen as well as cellular proteins including actin and tubulin were identified (Fu et al. 2005).
Comparison with existing literature revealed that 26 of the 35 identified albumin-associated proteins have been previously reported potential biomarkers (Anderson 2005; Polanski and Anderson 2006). Of these, nine were observed only in the albumin-enriched fraction and not in the albumin-depleted fraction (Sheng et al. 2006). Proteins in this category are α-2HS-glycoprotein, apolipoprotein A-I, ceruloplasmin, inter-α-trypsin inhibitor H4, kininogen, apolipoprotein C-III, fibrinogen, retinol-binding protein, and β-thromboglobulin. Furthermore, five of the nine albumin-bound potential biomarkers were observed only in the albumin-enriched fraction (α-2HS-glycoprotein, apolipoprotein A-I, apolipoprotein C-III, retinol-binding protein, and ceruloplasmin) and were observed in intact form. Since intact potential biomarkers were observed exclusively in the albumin-enriched fraction, therefore it is crucial to know in which fraction a potential biomarker will be located after sample fractionation with the caveat being whether all proteins present in the original sample will be represented in the depleted fraction. This makes the albumin-enriched fraction, and particularly the albuminome, an important sub-proteome in biomarker discovery.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree