(1)
Institut Bergonié, Bordeaux, France
Abstract
The Hedgehog pathway is an important signalling pathway for the development of vertebrates and invertebrates. It is involved in embryogenesis and morphogenesis and plays a major role in the renewal and differentiation of stem cells. Ligands and receptors are normally not expressed beyond embryonic life, except in stem cells. Loss of function of the actors of this pathway determines hereditary malformations, and activating mutations or gene overexpression is found in various malignancies.
Protein signals are issued in the vicinity of target cells equipped with adequate receptors. The ligands, first identified in Drosophila and called hedgehog (HHG) (by reference to the Drosophila larvae when affected by invalidating mutations of this gene), have been afterwards found in mammals. The membrane receptor of these ligands is called patched (PTCH). Receptor activation by the ligand induces the removal of a constitutive inhibition exerted by this receptor on a membrane protein called smoothened (SMO). This is followed by a cascade of events leading to the activation of transcription factors called GLI (for glial cells) and consequently the transcription of target genes required for proliferation.
9.1 Hedgehog Ligands
Three Hedgehog proteins are found in mammals; these are sonic hedgehog (gene SHH), Indian hedgehog (gene IHH) and desert hedgehog (gene DHH). They are first synthesised as a 45 kDa precursor, which undergoes an autocatalytic cleavage releasing a 19 kDa N-terminal peptide involved in signalling and a 45 kDa C-terminal peptide, which is involved in cleavage and bears a catalytic activity of cholesterol transferase.
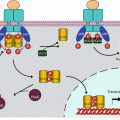
HHG proteins are subjected to post-translational modifications that are required for their activity; on the C-terminal side, they are covalently bound to a cholesterol molecule; on the N-terminal side, a specific palmitoyltransferase, HHAT (hedgehog acyltransferase) or SKI (skinny hedgehog), binds them to a palmitic acid moiety at the level of a cysteine residue. When equipped with these two hydrophobic ligands, HHG proteins can be inserted in the peripheral phospholipid layer of a lipoprotein and thus form a multimeric complex (Fig. 9.1).
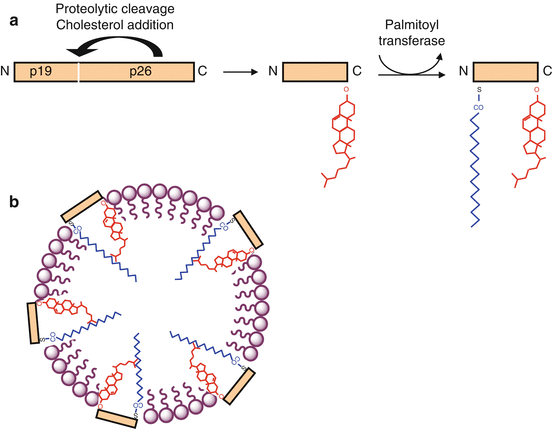

Fig. 9.1
HHG ligands and their post-translational modifications. (a) Native HHG proteins (45 kDa) are autocatalytically cleaved to generate a signalling fragment, which is covalently bound to a cholesterol molecule on its C-terminal serine. Afterwards, a palmitoyltransferase HHAT adds a palmitic acid to a cysteine residue located at the N-terminal extremity. (b) The molecule, with two lipid substituents, is inserted in phospholipid micelles, as part of lipoprotein structures
The mechanism of secretion of the HHG ligands is not precisely known. It involves a 12-transmembrane-domain protein called dispatched (DISP), which is indispensable and behaves as a membrane transporter of HHG proteins. This protein contains a sterol-binding domain, which should intervene in the processes of recognition and secretion of the HHG proteins.
9.2 Patched Receptors and Their Activation
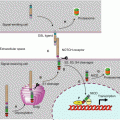
The HHG ligands with their two lipid substituents are recognised on a target cell by a receptor called patched (PTCH). There are two distinct PTCH proteins, encoded by the genes PTCH1 and PTCH2, which seem to have different functions, the first one being the best known and the only one to be considered here. PTCH belongs to the same transporter family as DISP and also presents 12 transmembrane domains and a cholesterol-binding domain. It also presents two large extracellular domains, which ensure HHG binding. PTCH is only an intermediary in HHG signal reception, since it transfers the information thus received to another membrane protein called smoothened (gene SMO), by releasing the constitutive inhibition exerted in this protein. The mechanism of this release is not intimately known, but it seems that no direct interaction exists between PTCH and SMO. PTCH would only be a transport protein and would control the localisation of a small inhibitory or excitatory molecule that remains to be identified. In the absence of signal, PTCH would pump an SMO inhibitory molecule from the inside to the outside of the cell; when bound to HHG, PTCH would cease pumping and SMO would then be functional; the reverse would be true if the molecule could conversely activate SMO (Fig. 9.2). Some authors hypothesise that the small molecule transported by PTCH and interacting with SMO could be vitamin D3 or an oxysterol.
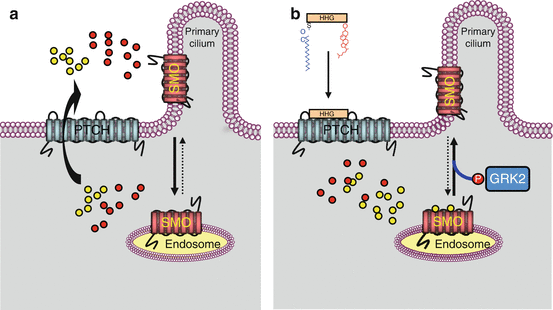

Fig. 9.2
PTCH receptor information transduction to SMO proteins. This figure presents a possible model for SMO activation by the PTCH receptor following HHG ligand binding. The SMO membrane protein is inactive in its endosomal localisation and is activated after migration to the primary cilium. In a first hypothesis, a substrate of PTCH transport activity (oxysterol?) would favour SMO migration to primary cilia and stabilise the active form of SMO. In the absence of HHG ligand (schema a), PTCH would pump oxysterol outside the cell, preventing thus its positive action on SMO ciliary localisation. In the presence of the HHG ligand (schema b), PTCH pumping activity would be inhibited, and the oxysterol would be available for maintaining SMO under its active form. In a second hypothesis, the substrate of the transport activity of PTCH (vitamin D3?) would be an inhibitor of SMO and would favour its inactive, endosomal localisation. In the absence of HHG ligand (schema a), PTCH would pump this sterolic molecule outside the cell where they would inhibit SMO. In the presence of HHG ligands (schema b), PTCH pumping activity would be inhibited, and SMO would no longer be destabilised by the inhibitory sterol. The two hypotheses are presented on the same schemas, the putative SMO activator (oxysterol?) as yellow circles and the putative SMO inhibitor (vitamin D3?) by red circles
SMO is a membrane protein with seven transmembrane domains, belonging to the fifth family of GPCRs (G-protein-coupled receptors) (Chap. 6) as FZD receptors (Chap. 7). When PTCH binds an HHG ligand, SMO accumulates in primary cilia, which are subcellular organelles present in all vertebrate cells and form protrusions at the surface of the plasma membrane, devoid of motility and based upon a microtubule cytoskeleton. SMO migration to primary cilia is required for Hedgehog signalling. This migration is accompanied by SMO phosphorylation by GRK2 (GPCR kinase 2, gene ADRBK1), which is thought to be required for migration. Proteins other than PTCH are able to bind HHG ligands and modulate the signal brought to the target cell: CDO (cell adhesion molecule-related/downregulated by oncogenes) and BOC (brother of CDO), which are members of the immunoglobulin superfamily and function as accessory receptors to HHG; it is not clear whether or not they are required for signal transmission. They operate via a glycosaminoglycan (heparan sulphate)-binding site. Another protein, HHIP (hedgehog-interacting protein), would be a decoy receptor for HHG ligands and would thus inhibit this signalling pathway.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree