10
Evolution of Recognition Molecules: The Immunoglobulin Superfamily
At this point it may be worth re-emphasizing the difference between ‘innate’ and ‘adaptive’ immunity, which lies essentially in the degree of discrimination of the respective recognition systems.
Innate immune recognition, e.g. by phagocytic cells, NK cells or the alternative complement pathway, uses a limited number of different receptors (more are being discovered all the time, but there are probably only a few dozen in total), which have evolved to recognize directly the most important classes of pathogen (see Figs 3 and 5).
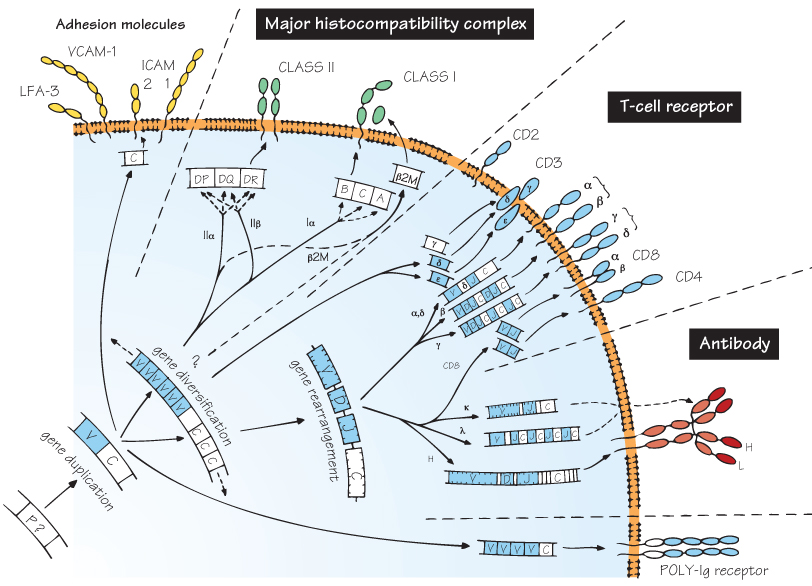
Recognition by lymphocytes, the fundamental cells of adaptive immunity, is quite another matter. An enormous range of foreign substances can be individually distinguished and the appropriate response set in motion. This is only possible because of the evolution of three sets of cell-surface receptors, each showing extensive heterogeneity, namely the antibody molecule, the T-cell receptor and the molecules of the major histocompatibility complex (MHC). Thanks to molecular biology, the fascinating discovery was made that all these receptors share enough sequences, at both the gene (DNA) and protein (amino acid) level, to make it clear that they have evolved from a single precursor, presumably a primitive recognition molecule of some kind (see Figs 3 and 46). The three-dimensional structure of all these receptors which was obtained more recently using X-ray crystallography has confirmed this close relationship.
Because antibody was the first of these genetic systems to be identified, they are often collectively referred to as the immunoglobulin gene superfamily, which contains other related molecules too, some with immunological functions, some without. What they all share is a structure based on a number of folded sequences about 110 amino acids long and featuring β-pleated sheets, called domains (shown in the figure as oval loops protruding from the cell membrane).
Much work is still needed to fill in the evolutionary gaps, and the figure can only give an impression of what the relationships between this remarkable family of molecules may have been. Their present-day structure and function are considered in more detail in the following four figures.
P?
The precursor gene from which the immunoglobulin superfamily is presumed to have evolved. It is believed that the key to the evolutionary success of the characteristic immunoglobulin domain is its extreme resistance to chemical or physical destruction. The gene has not been identified in any existing species, but may well have coded for a molecule that mediated cell–cell recognition. Alternative mechanisms for generating very diverse families of recognition molecules have been discovered very recently in several invertebrates and primitive vertebrates, some of which seem to be based on the leucine-rich repeat (LRR) protein domain instead of the immunoglobulin domain (see Fig. 5).
V, C
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree