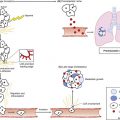
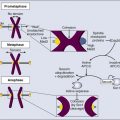
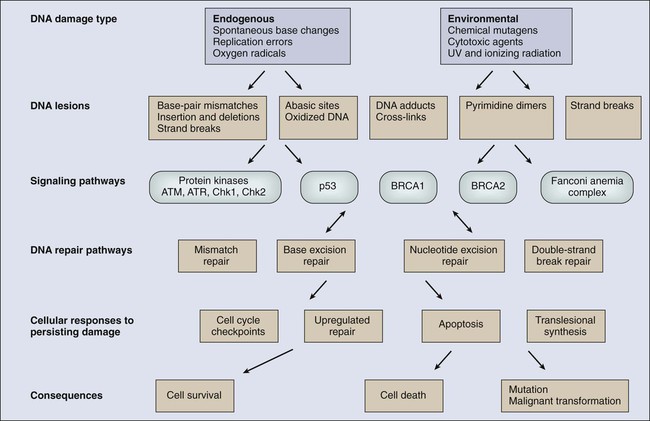
James M. Ford and Michael B. Kastan • DNA repair and the cellular response to DNA damage are critical for maintaining genomic stability. • Defects in DNA repair or the response to DNA damage encountered from endogenous or external sources results in an increased rate of genetic mutations, often leading to the development of cancer. • Inherited mutations in DNA damage response pathway genes often result in cancer susceptibility. • The major active pathways for DNA repair in humans are nucleotide excision repair, base excision repair, mismatch DNA repair, translesional DNA synthesis, and homologous recombination or nonhomologous end joining processes for double-strand break repair. • Inherited defects in nucleotide excision repair lead to the skin cancer–prone syndrome xeroderma pigmentosum, as well as Cockayne syndrome and trichothiodystrophy. • Inherited defects in base excision repair can result in colorectal cancer susceptibility. • Inherited defects in mismatch repair result in Lynch syndrome (hereditary nonpolyposis colorectal cancer syndrome), leading to increased incidence of gastrointestinal cancers, endometrial cancer, and other malignancies. • Inherited defects in DNA double-strand break repair and response pathways underlie a number of cancer prone disorders, including ataxia-telangiectasia, Nijmegen breakage syndrome, Bloom syndrome, Werner syndrome, Rothmund-Thomson syndrome, and Fanconi anemia. • Persons with Li-Fraumeni syndrome, who are highly prone to cancer because of inherited p53 mutations, and persons with breast-ovarian cancer syndrome, who are highly prone to cancer because of inherited mutations of the BRCA1 and BRCA2 genes, exhibit defects in multiple DNA repair and DNA damage response pathways. • Because most cancer therapeutic agents that are currently used damage DNA, understanding how normal cells and tumor cells respond to and repair DNA damage is an important aspect of understanding cancer therapeutic responses and toxicities of treatment. • The presence of mutations in DNA damage response and repair pathways in tumors present opportunities for developing therapeutics that target alternative damage response pathways and can be selectively lethal to the mutated tumor cells, an approach called “synthetic lethality.” Cancer is a disease caused in large part by the accumulation over time of changes to the normal DNA sequence, resulting in alterations, loss, or amplification of genes important for normal cellular functions and growth properties, including many proto-oncogenes and tumor suppressor genes. Nearly all cancers are clonal in origin; that is, they originate from a single progenitor cell rather than from a group of cells. The development of cancer in a particular cell type or tissue is caused by a series of specific mutations, each of which could be caused by DNA replication errors or unrepaired endogenous or exogenous DNA damage, or which could be the result of inherited mutations. In persons with the most common cancers, multiple genetic events occur in many different genes during the process of carcinogenesis, suggesting that an early and perhaps necessary event in the cancer process is an underlying defect in mechanisms to maintain genomic stability.3–3 In fact, alterations in the specific genes required for recognizing, processing, and responding to DNA damage may result in an enhanced rate of accumulation of additional mutations, recombinational events, chromosomal abnormalities, and gene amplification.4 Whether such a “mutator phenotype” is a required event for human tumorigenesis remains a controversial issue,5,6 but clear examples of such a process are evident and will be considered here. Many converging lines of experimental evidence reveal the complexity of the cellular responses to DNA damage and their role in malignant transformation.7 A number of interrelated biochemical pathways exist that influence the following actions: (1) the metabolism of potentially mutagenic or carcinogenic agents, (2) the efficiency and manner by which damaged DNA is recognized and repaired, (3) cell cycle progression and the coordination of DNA replication and cell division relative to the repair of lesions, and (4) the decision point determining survival or the active induction of programmed death of cells carrying different types and amounts of DNA damage. Many cellular pathways have evolved that require hundreds of gene products for the repair of DNA damage and involve excision of damaged DNA bases, joining of broken DNA strands, or replication bypass of DNA lesions (Table 10-1). The central role of DNA damage responses in neoplastic transformation has been highlighted by the discovery that mutations in numerous classes of genes required for DNA repair and the maintenance of genomic integrity result in a predisposition to the development of many malignancies.8 In fact, a number of rare, inherited disorders have been described that appear to be caused by defects in the repair of DNA lesions (Table 10-2), and many of these disorders are associated with an increased risk of the development of cancers.9 Table 10-1 Table 10-2 Human Genetic Diseases Involving Defects in DNA Damage Response Pathways DNA undergoes many types of spontaneous modifications, and it also can react with physical and chemical agents, some of which are endogenous products of normal cellular metabolism (e.g., reactive oxygen species), whereas others, including ionizing radiation and ultraviolet (UV) light, are threats from the external environment (Fig. 10-1). One pronounced example is exposure to genotoxic compounds in cigarette smoke, which currently are responsible for the most common cancers in Western countries. Most active chemotherapeutic agents function by damaging DNA through alkylation, cross-linking, and other means, and mechanisms to repair these lesions determine the sensitivity and resultant resistance of a tumor to such treatments. Damage to DNA can cause genetic mutations, and these mutations can lead to the development of cancer. DNA damage also may result in cell death that can have serious consequences for the organism of which the cell is a part—for example, loss of irreplaceable neurons in the brain. Accumulation of damaged DNA is thought to contribute to some of the features of aging.8 Therefore it is not surprising that a complex set of cellular surveillance and repair mechanisms has evolved to reverse or limit potentially deleterious DNA damage. Some of these DNA repair systems are so important that life cannot be sustained without them. An increasing number of human hereditary diseases that are characterized by severe developmental problems or a predisposition to cancer have been linked to deficiencies in DNA repair (Table 10-2). The results of DNA damage are diverse and usually adverse. Acute effects arise from disturbed DNA metabolism, triggering cell-cycle arrest or cell death. Long-term effects result from irreversible mutations contributing to oncogenesis and inherited genetic disorders. Many lesions block transcription, which has elicited the development of a dedicated repair system, transcription-coupled repair (TCR), which displaces or removes the stalled RNA polymerase and ensures preferential repair of lesions within the transcribed strand of expressed genes.12–12 Transcriptional stress due to DNA lesions that block RNA polymerase and DNA strand breaks caused by DNA damage or stalled replication forks constitute two major signals for DNA damage–inducible responses, including apoptosis,15–15 through both p53-dependent and independent mechanisms.16 Lesions also may interfere with DNA replication. A class of more than 10 specialized DNA polymerases were discovered that appear devoted to overcoming damage-induced replication stress.19–19 These special polymerases take over temporarily from the stalled replicative DNA polymerases. Although translesion polymerases protect the genome, this solution to replication blocks comes at the expense of a higher replicative error rate, and mutations in some of these polymerases cause cancer susceptiblity.9 Therefore detection of DNA lesions may occur by blocked transcription, replication, or specialized sensors. DNA damage checkpoints initially were defined as regulatory pathways that control the ability of cells to arrest the cell cycle in response to DNA damage, allowing time for repair.20 However, in addition to controlling cell cycle arrest, proteins involved in these pathways have been shown to control the activation of DNA repair pathways,21–25 the movement of DNA repair proteins to sites of DNA damage,26–31 and activation of transcriptional responses.34–34 When damage is too significant, a cell may opt for the ultimate mode of rescue by initiating apoptosis to benefit the organism at the expense of losing a cell.37–37 Because the DNA damage response pathway has been better defined at a molecular level, it has been seen as a network of interacting pathways that together execute the response.38 Initial recognition of DNA damage occurs by a variety of damage-specific DNA binding proteins; these proteins, either by themselves or together with complexes of associated proteins that are not directly involved in DNA repair, may signal the DNA damage response.39 Transduction and amplification of the DNA damage signal often is carried out by an overlapping set of conserved protein kinases, including the phosphoinositide-3-kinase-related proteins, which include ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) proteins, the checkpoint kinases Chk1 and Chk2, and others.7,40–45 Many of these protein kinases are themselves targets for phosphorylation and activation; they then further target downstream genes critical to oncogenesis, such as p53 and BRCA1.40,46–48 The ultimate targets of this highly regulated DNA damage response include mechanisms for DNA repair, and although much of DNA repair is constitutive, a number of regulatory connections between the DNA damage response pathway and DNA repair have emerged.7 In mammals, a large number of genes involved in DNA repair are transcriptionally induced in response to DNA damage, suggesting that many facets of repair are inducible, similar to the RecA-dependent SOS response in bacteria that enhances DNA repair and mutagenesis after DNA damage.25,49 In fact, the p53 tumor suppressor gene is a central mediator of the DNA damage–inducible transcriptional response in humans, and p53 mutant mammalian cells are deficient in several aspects of DNA repair.21–25,34,50 Therefore the mammalian DNA damage–inducible response pathway is highly regulated and fine-tuned to determine if a particular cell type proceeds to a cell cycle checkpoint and DNA repair, or cell death, after an insult with significant damage. Defects at any level of these pathways can alter repair and result in carcinogenesis (Fig. 10-1). DNA repair may be defined as those cellular responses associated with the restoration of the normal nucleotide sequence after events that damage or alter the genome.51 Given the wide variety of DNA damage a cell encounters, it is not surprising that an equally large number of repair systems are available to handle these insults. Indeed, many of the repair systems are broadly overlapping and interacting, with several sharing certain strategies and even specific gene products. Much of what is known regarding the basic mechanisms of many types of DNA repair comes from the study of lower organisms, such as bacteria and yeast, because these pathways have been highly conserved through evolution. Inherited defects in any of the major DNA repair pathways in humans, in general, predisposes to malignancy, and several of these syndromes will be discussed in detail. In humans, a great deal has been learned regarding DNA repair from the often rare, autosomal-recessive hereditary syndromes associated with defects in DNA repair genes.9 The most versatile and ubiquitous mechanisms for DNA repair are those in which the damaged or incorrect part of a DNA strand is excised and then the resulting gap is filled by repair replication using the complementary strand as a template. The redundancy of genetic information provided by the duplex DNA structure is essential to the maintenance of the genome by this “cut and patch” mode known as excision repair. Each DNA strand can serve as a template for replication-based repair of the other strand. Excision repair was discovered in the early 1960s through basic studies on the effects of UV irradiation on DNA synthesis and repair replication in bacteria.54–54 Nucleotide excision repair (NER) functions to remove many types of lesions, including bulky base adducts of chemical carcinogens, intrastrand cross-links, and UV-induced cyclobutane pyrimidine dimers and 6-4 photoproducts. Such lesions may serve as structural blocks to transcription and replication as a result of distortion of the helical conformation of DNA, and they also may result in mutations if translesional replication occurs or if they are not repaired correctly. The sequential steps for NER are (1) recognition of the damaged site, (2) incision of the damaged DNA strand near the site of the defect, (3) removal of a stretch of the affected strand containing the lesion, (4) repair replication to replace the excised region with a corresponding stretch of normal nucleotides using the complementary strand as a template, and (5) ligation to join the repair patch at its 3′ end to the contiguous parental DNA strand.55,56 This excision repair pathway can remove DNA damage from sites throughout the genome and is termed global genomic repair (GGR). Most human NER genes have been identified and cloned, and many have been shown to be mutated in persons with hereditary NER-deficient, cancer-prone diseases (Fig. 10-2).25,50,57 A unique problem arises if a bulky lesion is encountered by a translocating RNA polymerase-making messenger RNA before repair enzymes have removed the damage and restored intact DNA. The polymerase may be arrested at the site of the lesion and prevent access to the damage by repair enzymes. Furthermore, the arrest of transcription in human cells is a strong signal for p53 activation and can trigger apoptosis.16 In this situation, a dedicated excision repair pathway known as transcription-coupled repair (TCR) comes to the rescue to displace the RNA polymerase and then efficiently repairs the blocking lesion so that transcription may resume and the cell may survive.49 The role of defects in TCR, if any, in cancer development remain unclear. Recent evidence suggests that cells from patients with Cockayne syndrome (CS), trichothiodystrophy (TTD), and xeroderma pigmentosum (XP)/CS share a defect in repair of oxidative DNA damage, which may explain the overlapping progeroid features of these syndromes.58 Recently it has become apparent that the GGR subpathway of NER is damage inducible and highly regulated by both transcriptional and posttranslational mechanisms after DNA damage, in concert with damage-inducible cell cycle checkpoints and apoptosis.7,50 In fact, the p53 gene, which is central to maintaining genomic stability in human cells, is required for efficient GGR of UV light–induced and carcinogen-induced DNA damage and functions as a DNA damage–activated transcription factor that directly regulates the expression of several NER damage recognition genes.23–25,34 Similarly, several other important cancer-related genes have been shown to transcriptionally regulate the DNA damage recognition NER genes XPC and DDB2, including BRCA1 and E2F1.61–61 Therefore the GGR pathway of NER appears relevant to suppressing DNA damage–induced malignancy and is highly regulated by genes involved in tumor suppression. Further evidence for the importance of exquisite regulation of DNA damage recognition and repair activity to carcinogenesis comes from a new appreciation for the role of the ubiquitin-proteasome system in maintenance of genomic stability.62 Many complex intracellular signaling processes, including DNA repair, are controlled not only by the regulated expression of proteins but also by their assembly and targeted degradation through posttranslational ubiquitination. With regard to NER, the same DNA damage recognition proteins found to be transcriptionally induced after DNA damage (XPC and DDB2) are also rapidly ubiquitylated after DNA damage by an E3 ubiquitin ligase activity that contains the DDB2 binding partner DDB1, resulting in a higher order of regulation.50 The ubiquitin-proteasome system has been found to regulate other repair pathways as well, including translesional synthesis and the Fanconi anemia–associated homologous recombination.62 Therefore mammalian cells have evolved a proteolytic pathway to limit their repair capability through restricting certain types of DNA repair activities. Considering that most DNA damage recognition complexes identify a variety of DNA damages or metabolic conditions rather than binding to specific DNA sequences, it is plausible that a “checkpoint” mechanism is required to limit DNA damage binding from interfering with other cellular processes that involve unconventional DNA structures, and that after inducible expression of DNA repair genes, levels are actively reduced to avoid gratuitous DNA repair and associated mutagenesis mediated by error-prone polymerases. Despite the plethora of mechanisms for DNA repair described herein, it is likely that the cellular replication machinery will nevertheless encounter lesions in the DNA template strand that block replication during each cell cycle. The solution adapted by cells is DNA damage tolerance, in which DNA is synthesized past the damaged bases and subsequently excised after the replication fork has passed—a process known as translesional synthesis and carried out by a newly identified class of specialized error-prone DNA polymerases, termed ζ (zeta) to σ (sigma).19–19 These Y-family DNA polymerases take over temporarily from the stalled replicative DNA polymerases (δ [delta] and ε [epsilon]). They have more flexible base-pairing properties, thus permitting translesion DNA synthesis, with each polymerase probably designed for a specific category of injury. Although translesion polymerases protect the genome, this solution to replication blocks comes at the expense of a higher error rate. For instance, inherited defects in polymerase eta (pol η), encoded for by the XPV/POLH/RAD30 gene, which specializes in relatively error-free bypassing of UV-induced cyclobutane pyrimidine dimers, causes a variant form of the skin cancer–prone disorder XP.63,64 A direct correlation between unrepaired DNA damage and carcinogenesis in humans was first established when James Cleaver found that the cancer-prone hereditary disease XP involved a defect in the repair of DNA lesions produced by UV light.65 Since then, at least four syndromes have been attributed to inborn errors in NER: XP, CS, TTD, and UV sensitivity syndrome (UVSS), all of which are characterized by exquisite sun sensitivity. XP is a rare, autosomal-recessive disease in which homozygous individuals display several characteristics: (1) extreme sensitivity of the skin to sun exposure, which is evident by 1 year of age; (2) pigmentation abnormalities and premalignant lesions in sun-exposed skin; (3) increases up to 4000-fold in the incidence of skin cancers (predominantly squamous and basal cell carcinomas, but also melanomas) and ocular neoplasms, occurring three to five decades earlier than in the general population; and (4) a ten- to twentyfold increased incidence of internal cancers in non—sun-exposed sites.9,66,67 Overall, life span is reduced by approximately 30 years among persons with XP, and many die as a result of malignancies.68 Approximately 20% of patients with XP also display progressive neurological degeneration, characterized by peripheral neuropathy, sensorineural deafness, progressive mental retardation, and cerebellar and pyramidal tract involvement.69 XP occurs worldwide in all ethnic groups and with a frequency varying from 1 to 10 patients per million. The biochemical defect in cells in most persons with XP is in NER,65 although in a small number of cases (termed XP variants), excision repair appears normal and a defect exists in bypass replication at unrepaired lesions as a result of a mutation in the pol η translesional synthesis gene (XPV).70 Complementation analysis via fusion of cells from different patients has demonstrated genetic heterogeneity within XP and provided evidence for the existence of at least seven excision-deficient complementation groups, termed XP-A to XP-G, in addition to XP variant.9 CS is an autosomal-recessive disease that is associated with defective TCR of UV-damaged and oxidative-damaged DNA.73–73 It is characterized by cutaneous photosensitivity, cachectic dwarfism, skeletal abnormalities, retinal degeneration, cataracts, severe mental retardation, and neurological degeneration characterized by primary demyelination.69,74 In contrast to patients with XP, those with CS are not at increased risk for the development of skin cancers. The average life span of persons with CS is only 12 years, with most patients succumbing to infectious or renal complications, rather than cancer.75 CS is characterized by the existence of at least three complementation groups. Several patients in XP groups B, D, and G have been found to share the DNA repair defects and clinical features of CS together with the cutaneous manifestations of XP.76,77 One model for the severe developmental problems in persons with CS is that endogenous oxidative damage in metabolically active cells, including neurons, is blocking transcription. The lack of TCR could then lead to apoptosis of these essential cells. Alternatively, CS could be a “transcription disease” caused by defects in TFIIH-associated gene products and resulting in inadequate gene expression. TTD is an autosomal-recessive condition sharing many of the signs and symptoms of CS, with the additional hallmark of brittle, sulfur-deficient hair and nails due to reduced cysteine content in the component proteins. As with CS, several of the responsible genes implicated are XPB and XPD, but there is a third complementation group, TTD-A, in which mutations of a small 8 kDa TFIIH stabilizing factor GRF2H5 have been identified.58 The favored model for TTD is that of a transcription deficiency with respect to the genes relevant to the phenotype, including sulfur-containing proteins.78 It is also conceivable that TTD and CS could be diseases of “premature cell death” in which the transcription deficiency and deficiency in TCR could cause the apoptosis of certain classes of metabolically active cells that sustain significant endogenous oxidative damage (e.g., neurons). UVSS is a rare disease characterized by severe photosensitivity but without cancer susceptibility. As with CS, cells from the three identified complementation groups lack TCR. Through use of exome sequencing and genetic and proteomic approaches, authors of three recent studies have identified mutations in the UV-stimulated scaffold protein A (UVSSA) gene as being responsible for UVSS group A. These findings suggest a new mechanistic model involving UVSSA and the ubiquitin-specific protease 7 (USP7) in the fate of stalled RNA polymerase II during TCR.79 Analysis of the specific abnormalities in NER displayed by the various genetic complementation groups of XP, CS, TTD, and UVSS allow correlations to be drawn with their heterogeneous clinical features. Specifically, only the subgroups of patients who display a defect in GGR are at significantly increased risk for developing UV-induced malignancies. In contrast, the neurological symptoms and developmental abnormalities associated with other complementation groups of XP and CS are found only in the groups that are defective in TCR. The fact that the TFIIH complex, containing the XP-B and XP-D proteins, is common to both core NER and transcriptional initiation, supports the suggestion that the clinical phenotype of patients with defects in TCR may actually be due to abnormalities in transcription, rather than in repair itself.78 Although these observations may explain the molecular basis for many of the clinical characteristics of XP and CS, they present an apparent paradox with regard to the cancer risk of these patients. Many currently recognized oncogenes and tumor suppressor genes are known to possess important cellular functions and to be actively expressed in normal cells. Because CS cells are defective in the repair of actively expressed genes, it would be reasonable to expect that patients with CS would acquire mutations in genes leading to transformation more readily than normal patients. However, this expectation is not supported by the clinical picture. It has been demonstrated that defects in TCR specifically activate DNA damage–induced apoptosis,13,14 which may eliminate potentially mutagenic premalignant cells. A major source of DNA damage to cellular genomes arises from normal metabolism in the cytoplasmic environment through hydrolysis and exposure to reactive metabolites that cause oxidation and alkylation of DNA. The repair system primarily involved in identifying and removing such lesions, as well as for dealing with the spontaneous loss of purines from DNA, is the base excision repair (BER) pathway.80,81 The essential nature of BER for viability is highlighted by the fact that although a number of BER proteins have been discovered, only recently has a single human hereditary disease been identified that appears to result from a mutation in a gene unique to this pathway.82–85 The enormous task required for BER is exemplified by the fact that in 1 hour, humans spontaneously lose on the order of a trillion guanines from their DNA, each of which must be replaced. Similarly, a large number of cytosines become deaminated spontaneously and the resulting product, uracil, must be removed and replaced with cytosine to restore the correct nucleotide sequence. In most cases, BER is initiated by one of a set of lesion-specific glycosylases that recognize the altered or inappropriate base and cleaves it from its sugar moiety in the DNA (Fig. 10-3
DNA Damage Response Pathways and Cancer
Introduction
DNA Repair Pathway
Type of DNA Damage
Approximate No. of Genes
Nucleotide excision repair
Bulky or helix-distorting DNA adducts, for example, ultraviolet photoproducts and carcinogen adducts
37
Base excision repair
Oxidative DNA damage; spontaneous depurination
40
Mismatch repair
Mispaired nucleotides
1-15 nucleotide insertion-deletion loops
26
Homologous recombination
Double-strand DNA breaks, DNA cross-links
20
Nonhomologous end joining
Double-strand DNA breaks
10
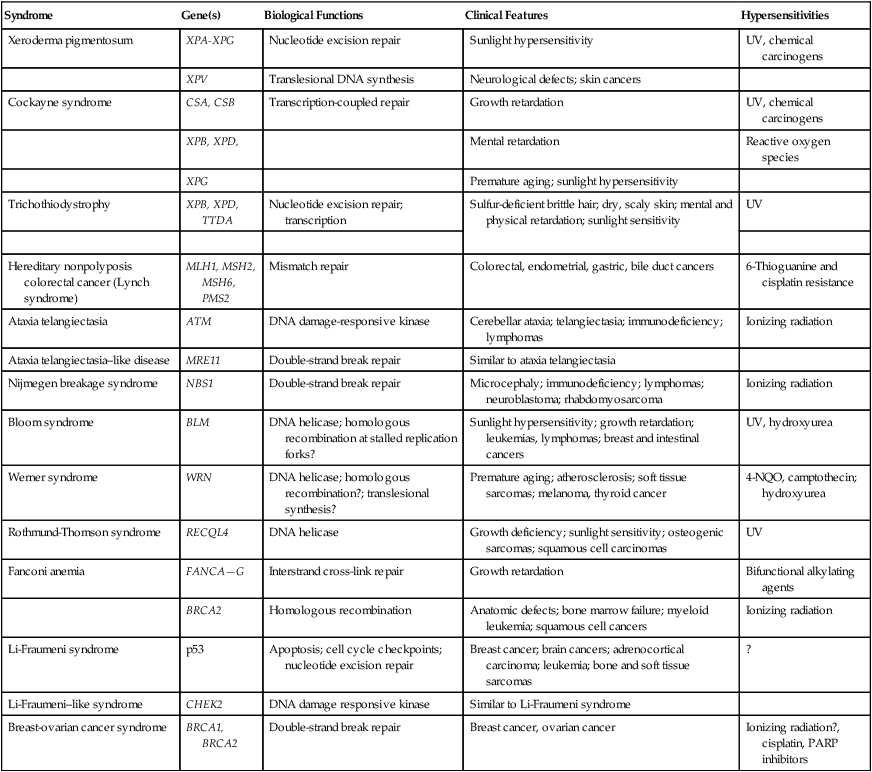
Syndrome
Gene(s)
Biological Functions
Clinical Features
Hypersensitivities
Xeroderma pigmentosum
XPA-XPG
Nucleotide excision repair
Sunlight hypersensitivity
UV, chemical carcinogens
XPV
Translesional DNA synthesis
Neurological defects; skin cancers
Cockayne syndrome
CSA, CSB
Transcription-coupled repair
Growth retardation
UV, chemical carcinogens
XPB, XPD,
Mental retardation
Reactive oxygen species
XPG
Premature aging; sunlight hypersensitivity
Trichothiodystrophy
XPB, XPD, TTDA
Nucleotide excision repair; transcription
Sulfur-deficient brittle hair; dry, scaly skin; mental and physical retardation; sunlight sensitivity
UV
Hereditary nonpolyposis colorectal cancer (Lynch syndrome)
MLH1, MSH2, MSH6, PMS2
Mismatch repair
Colorectal, endometrial, gastric, bile duct cancers
6-Thioguanine and cisplatin resistance
Ataxia telangiectasia
ATM
DNA damage-responsive kinase
Cerebellar ataxia; telangiectasia; immunodeficiency; lymphomas
Ionizing radiation
Ataxia telangiectasia–like disease
MRE11
Double-strand break repair
Similar to ataxia telangiectasia
Nijmegen breakage syndrome
NBS1
Double-strand break repair
Microcephaly; immunodeficiency; lymphomas; neuroblastoma; rhabdomyosarcoma
Ionizing radiation
Bloom syndrome
BLM
DNA helicase; homologous recombination at stalled replication forks?
Sunlight hypersensitivity; growth retardation; leukemias, lymphomas; breast and intestinal cancers
UV, hydroxyurea
Werner syndrome
WRN
DNA helicase; homologous recombination?; translesional synthesis?
Premature aging; atherosclerosis; soft tissue sarcomas; melanoma, thyroid cancer
4-NQO, camptothecin; hydroxyurea
Rothmund-Thomson syndrome
RECQL4
DNA helicase
Growth deficiency; sunlight sensitivity; osteogenic sarcomas; squamous cell carcinomas
UV
Fanconi anemia
FANCA—G
Interstrand cross-link repair
Growth retardation
Bifunctional alkylating agents
BRCA2
Homologous recombination
Anatomic defects; bone marrow failure; myeloid leukemia; squamous cell cancers
Ionizing radiation
Li-Fraumeni syndrome
p53
Apoptosis; cell cycle checkpoints; nucleotide excision repair
Breast cancer; brain cancers; adrenocortical carcinoma; leukemia; bone and soft tissue sarcomas
?
Li-Fraumeni–like syndrome
CHEK2
DNA damage responsive kinase
Similar to Li-Fraumeni syndrome
Breast-ovarian cancer syndrome
BRCA1, BRCA2
Double-strand break repair
Breast cancer, ovarian cancer
Ionizing radiation?, cisplatin, PARP inhibitors
Types of DNA Damage
Consequences of DNA Damage
DNA Damage Response Pathways
Types of DNA Repair and Their Contribution to Cancer
Nucleotide Excision Repair
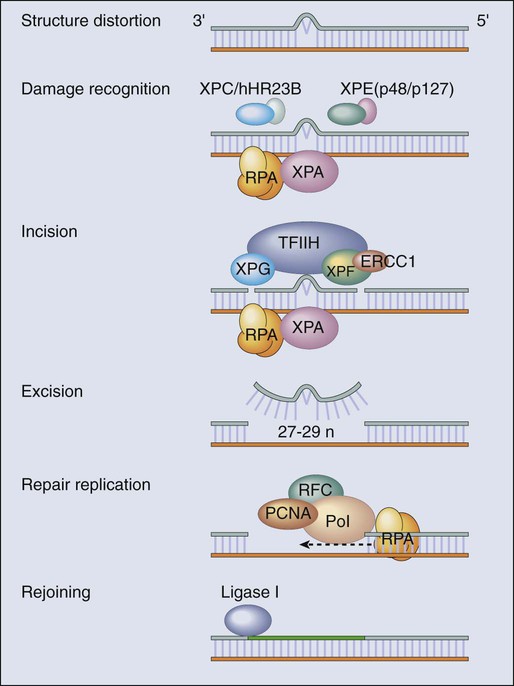
Human Nucleotide Excision Repair Deficient Syndromes and Cancer
Base Excision Repair
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


DNA Damage Response Pathways and Cancer