Fig. 1.1
Giemsa-stained normal metaphase spreads from bone marrow preparations of (a) mouse, showing all acrocentric chromosomes (2n = 40), and (b) rat, showing acrocentric, submetacentric, and metacentric chromosomes (2n = 42)
Mammalian species which are most commonly used in biomedical research are mouse, M. musculus (2n = 40); Norway rat, Rattus norvegicus (2n = 42); Syrian (Golden) hamster, Mesocricetus auratus (2n = 44); Chinese hamster, Cricetulus griseus (2n = 22); dog, Canis familiaris (2n = 78); rhesus macaque, Macaca mulatta (2n = 42); African green monkey, Cercopithecus aethiops (2n = 60); and lately the South American marsupial, Monodelphis domestica (2n = 18). The laboratory mouse (hence called mouse only) has a relatively short gestation period, a large litter size, and a well-worked out reproductive physiology, genetics, and embryology. It is also rather easy to manipulate the mouse genome in the laboratory. Due to these favorable characteristics and many others, the use of laboratory mouse has dominated in biomedical research for manipulating genomes, making knocking-in and knocking-out genes and transgenics; the mouse has been used as a model species for all mammals. Realistically, it would be erroneous to consider the mouse as a model species for their cytogenetic characteristics among mammals. There are close to 50 species belonging to the genus Mus described and some studied cytogenetically. We and others have studied chromosomes of several of these species including M. musculus, M. dunni, M. booduga, M. platythrix, M. fulvidiventris, M. caroli, M. cervicolor, M. cookii, M. shortridgei, M. pahari, and many others (Pathak and Stock 1974; Pathak 1976; Wurster-Hill et al. 1973; Markvong et al. 1975, 1976; Pathak and Hsu 1976; Hsu et al. 1978). The diploid chromosome numbers in the genus Mus vary between 2n = 18 and 44. Are M. musculus chromosomes similar in structure and morphology to the chromosomes of other species of the genus Mus? How close are mouse chromosomes to human chromosomes? Is the mouse genome the most relevant to the mammalian model genome? There are no satisfactory answers available to some of these questions. Although Giemsa (G-) banding patterns of X chromosomes in most mammalian species are very similar, their morphology varies from metacentric to acrocentric (Pathak and Stock 1974). On the other hand, the size and morphology of the mammalian Y chromosomes range from a mere minute dot in the opossum, Didelphis albiventris, and in some species of bats to a very large acrocentric, as in the field vole, Microtus agrestis, or biarmed, as in human (Pathak 1983). However, the chromosome banding patterns and morphology of their autosomes vary considerably with only a small number of chromosomes showing similar banding conservatism (Mascarello et al. 1974).
In this mini review, I would like to present the highlights of mammalian chromosomes in brief and compare them with that of the laboratory mouse. This brief mammalian cytogenetic review is meant for scientists who are not cytogeneticists but are involved in studying developmental and molecular cancer biology, gene regulation, telomere dynamics, animal model for cancer research, drug toxicity testing, anticancer drug development, genomics, proteomics, and all other aspects which do not include the chromosomes of the laboratory mouse.
1.2 Do Large Size Mammals Have Longer and High Number of Chromosomes?
The mammals vary considerably in over all body size of the organism ranging from the largest, as the aquatic mammal – whale – to the smallest, as shrew. In eutherian mammals the range of diploid chromosome numbers vary between 2n = 6 and 2n = 102. The lowest diploid chromosome number reported of all mammalian species is 2n = 6 in female and 7 in male of the Indian muntjac, also called barking deer (Muntiacus muntjak: order, Artiodactyla; family, Cervidae), as shown in Fig. 1.2. It was first studied and reported by Wurster and Benirschke (Wurster and Benirschke 1970). The highest diploid number of all studied species is 2n = 102 in a rodent, Tympanoctomys barrerae (order, Rodentia; family, Octodontidae) reported by Contreras and associates (Contreras et al. 1990). After studying the chromosome constitutions of the aardvark (Pathak et al. 1980), Orycteropus afer (order, Tubulidentata; family, Orycteropodidae), which is 2n = 20, to zebra, Equus grevyi (A to Z) with a diploid number of 46 (order, Perissodactyla; family, Equidae), we have come to the conclusion that the body size of the adult animals bears no relation to the length, diploid number, and morphology of their chromosomes. Whales are among the largest aquatic mammals, and elephants are the largest surviving land mammals known on earth. Yet, the fin whale (Arnason 1969) (Balaenoptera physalus: order, Cetacea; family, Balaenopteridae) has 2n = 44 and African elephant (Hungerford et al. 1966) (Loxodonta africana: order, Proboscidea; family, Elephantidae) has a diploid number of 2n = 56. Both these largest-sized living mammals have a mixture of meta-, submeta-, and acrocentric chromosome morphology. Such chromosome morphology is also the characteristic of giraffe with a diploid number of 2n = 30. However, the largest-sized chromosomes are present in the Indian muntjac and the smallest size in some species of old-world bats (Pathak 1990). A related deer, Chinese muntjac, Muntiacus reevesi (order, Artiodactyla; family, Cervidae) has 2n = 46, with all acrocentric chromosome morphology (Wurster and Benirschke 1967). All cats (family: Felidae), irrespective of their size, from the small house cat to the large Siberian tiger, have a diploid chromosome number of 2n = 38, except only two species having Robertsonian translocation (Hsu et al. 1963) with 2n = 36. All breeds of dog, irrespective of their body size and shape, have 2n = 78 chromosomes of which autosomes are acrocentric and the X and Y are biarmed (Pathak et al. 1982). All species of rhinoceros have a diploid chromosome number of 2n = 82 (atlas of mammalian chromosomes) (O’Brien et al. 2006). In different groups of bats again, the animal body size does not correlate with their diploid chromosome numbers. For example, the flying fox, Pteropus giganteus giganteus (Ray-Chaudhuri et al. 1968) (suborder, Megachiroptera; family, Pteropodidae), has a diploid number of 2n = 38, whereas the smallest-sized microchiropteran species, Pipistrellus mimus (Pathak and Sharma 1969) (family: Vespertilionidae), also has 2n = 38. From these differences in mammalian diploid number, we conclude that chromosome number, length, and morphology, individually or in combination, do not confer any correlation with the body size of adult animals.
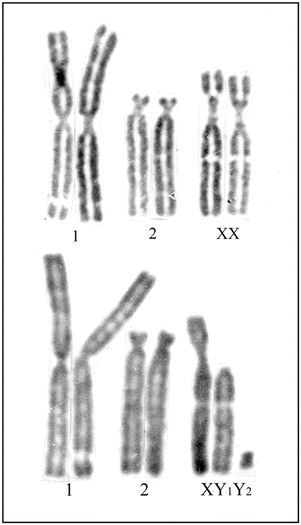

Fig. 1.2
Giemsa-stained karyotypes from normal fibroblast cultures of a female (upper row) and a male (lower row) of Indian muntjac showing X/autosome translocation. In this species, the female has 2n = 6 and male 2n = 7 chromosomes. The actual X chromosome is represented in the short of the third pair and the actual Y, the smallest biarmed chromosome (Y2)
1.3 Do X and Y Chromosomes Exist Intact in All Mammals?
In mammalian species, the sex-determining mechanisms are XX in female and XY in male, making a male, heterogametic. But in birds and reptiles, the female is heterogametic (ZW) and the male is homogametic (ZZ). As a rule, in all mammalian species including the marsupials, the Y chromosome is always smaller than the X chromosome (Galton 1966). Among all mammals, marsupials have generally the lowest diploid numbers. However, a few rare mammalian species have odd chromosome numbers within two sexes. For example, in Indian muntjac, the X chromosome is permanently fused onto an autosome, and therefore, the male has 2n = 7 and the female 2n = 6 chromosomes (Fig. 1.2). Similarly in Seba’s fruit bat, Carollia perspicillata (order, Chiroptera; family, Phyllostomidae) also, where the male has 2n = 21 and female 2n = 20, the X chromosome is permanently translocated to an autosome (Hsu et al. 1968; Pathak et al. 1973b). On the other hand in some rare mammalian species, the Y chromosome is also permanently translocated to an autosome. For example, in African and Indian mongoose, the male has 2n = 35, XO, and the female 2n = 36, XX chromosomes (Fredga 1965; Pathak and Stock 1976). At least in the African marsh mongoose, Atilax paludinosus (order, Carnivora; family, Viverridae), the Y chromosome is permanently translocated onto an acrocentric autosome (Pathak and Stock 1976).
I have previously reported in a book chapter all those mammalian species in which either X or Y chromosome is permanently fused to an autosome (Pathak 1983; see Tables I and II). The morphology and the banding patterns of the X and Y chromosomes in more than 90 mammalian species have also been published (Pathak and Stock 1974). The sex-determining mechanism in some rare rodent species is highly unusual. For example, in the mole vole, Ellobius lutescens (2n = 17), X0/X0, constitution exists in both sexes. In other species of Ellobius, the sex chromosomes are isomorphic in both genders (O’Brien et al. 2006; Chiarelli and Capanna 1973). In the creeping vole, Microtus oregoni, a unique chromosome pattern exits which is gonosomic mosaic where males have OY/XY chromosome constitution (Ohno et al. 1963).
1.4 Distribution of Constitutive Heterochromatin in Mammalian Species
The heterochromatin which is identified by a simple C-banding technique of Arrighi and Hsu (1971) has played an important role in mammalian speciation (Hsu and Arrighi 1971; Pathak et al. 1973a,c). The amount and distribution pattern of genomic heterochromatin directly correlate with the number of species present in given taxa of mammals. Rodents have the largest amount of heterochromatin and the largest number of species in which the diploid chromosome varies between 2n = 10 in Akodon sp. and 2n = 102 in Tympanoctomys barrerae. However, in the group Felidae, which includes small house and large jungle cats, the amount of heterochromatin is the smallest and only a limited number of species are known thriving in nature; mostly with 2n = 38 chromosomes, with the exception of only two species with a 2n = 36, due to a Robertsonian translocation (Hsu et al. 1963). Even the man’s best friend, the domestic dog is known to have the smallest amount of heterochromatin (Pathak et al. 1982). In the North American opossum, Didelphis virginiana (2n = 22), the heterochromatin is exclusively localized to the sex chromosomes and autosomes are almost devoid of any C-banding (Sinha et al. 1972). Order Chiroptera is the second largest, next to Rodentia group of mammals having the largest number of species and considerably large amount of genomic heterochromatin (Pathak et al. 1973b). Among different species of the genus Mus, the distribution of heterochromatin is mainly in the centromere regions except in M. dunni, where every autosome has a distinct heterochromatic short arm. In its biarmed X chromosome, the short (p) arm is totally heterochromatic, and a small amount of heterochromatin is present at the tip of its long (q) arm (Markvong et al. 1976; Pathak and Hsu 1976). In another Indian species, M. booduga (2n = 40), the entire p arms of some metacentric autosomes are heterochromatic (Sharma et al. 1990). Compared to the laboratory mouse, the amount of heterochromatin is much less in Rattus sp. The largest amount of heterochromatin reported among mammals is in the ground squirrels, Ammospermophilus (42 %) and Thomomys species (60 %) (Mascarello and Mazrimas 1977). Syrian hamster (2n = 44) and Chinese hamster (2n = 22) also have considerably large amount of genomic heterochromatin (Popescu and DiPaolo 1979; Arrighi et al. 1974). Most of these heterochromatic regions are telomeric DNA which will be discussed later on in this review.
In the human genome, each chromosome has C-band in the pericentromeric regions. However, chromosomes 1, 9, and 16 have larger blocks of C-banding compared to other human chromosomes. In the human Y chromosome, the distal end of the entire q arm is also heterochromatic (Fig. 1.3). The C-banding technique does not help in the identification of the entire length, but other procedures, such as Q, G, R, and SKY (Fig. 1.4), are used in the identification of chromosomes.
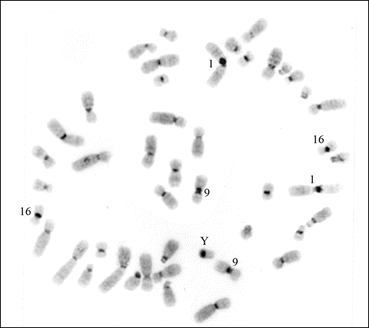
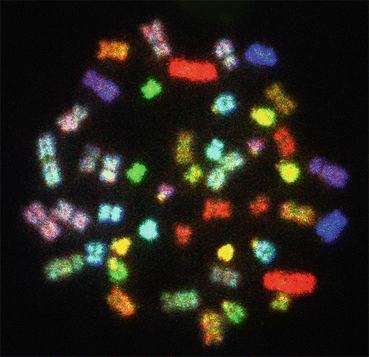

Fig. 1.3
A C-banded metaphase spread from the peripheral blood lymphocyte of the author showing the typical distribution of constitutive heterochromatin. The chromosomes 1, 9, 16, and the Y have more heterochromatin compared to other autosomes and the X chromosome
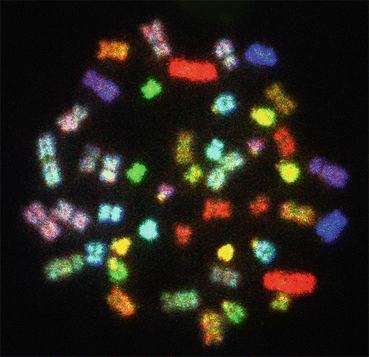
Fig. 1.4
A spectral karyotyping (SKY) image of a normal human female metaphase plate showing longitudinal differentiation
1.5 Locations of Ribosomal Cistrons in Mammalian Chromosomes
The chromosome banding techniques allow identification of individual chromosomes and, in some cases, provide information on the location of certain genes. The nucleolus organizer regions (NORs) that stain positively with the silver nitrate staining procedure are known to be the locations of ribosomal genes (Goodpasture and Bloom 1975). The NORs are generally positioned on the secondary constriction region of chromosomes whose number and location vary considerably among mammalian species. In the human genome, NORs are located in the short arms of ten acrocentric chromosomes belonging to the D and G groups (Fig. 1.5), whereas in most cats and in the rhesus monkey, only a pair of autosomes in each has interstitial NORs. In addition, considerable NOR polymorphism exists between homologous and nonhomologous chromosomes. In some mammalian species, the NORs are even located in the sex chromosomes as well as autosomes, as in the case of the South American opossum, Monodelphis domestica (2n = 18) (Merry et al. 1983). The location of NORs varies from proximal to distal ends (terminal) of the arms, as in the laboratory mouse and cattle chromosomes, respectively. In some other mammalian species, they are present in the middle of the arms.
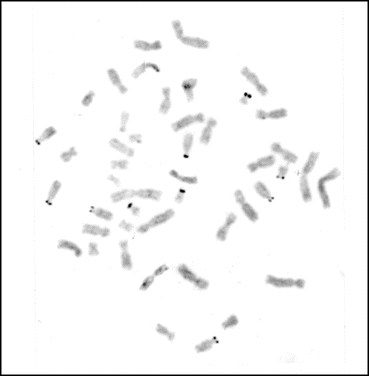

Fig. 1.5




A normal metaphase spread from the human peripheral blood lymphocyte showing the distribution of Ag-NORs in the short arms of D and G group chromosomes
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree