Highlights
- •
ctDNA provided complementary information on actionable mutations to tissue biopsy.
- •
ctDNA provided a comprehensive view of mutational landscape dynamics over time.
- •
ctDNA-based molecular relapse detection has high levels of clinical validity.
Abstract
Objectives
Head and neck squamous cell carcinoma (HNSCC) is characterized by significant genetic intra-tumor heterogeneity (ITH), which may hinder precision medicine strategies that depend on results from single tumor-biopsy specimens. Treatment response assessment relies on radiologic imaging, which cannot detect minimal residual disease (MRD). We assessed the relevance of circulating tumor DNA (ctDNA) as a biomarker for ITH and MRD in HNSCC.
Materials and methods
We recruited 41 non-metastatic resectable HNSCC patients treated with upfront curative-intent surgery in the prospective biobanking SCANDARE study (NCT03017573). Thirty-one patients (76 %) showed recurrent disease at a median follow-up of 41 months. Targeted next-generation sequencing was performed on resected tumor tissues, as well as on serial blood samples obtained at surgery, within 14 weeks after surgery, at six months and at recurrence.
Results
ctDNA was detected in 21/41 patients at surgery (sensitivity: 51 %; 95 % CI, 35–67 %) and 15/22 patients at recurrence (sensitivity: 68 %; 95 % confidence interval [CI], 45–86 %). Among patients with mutations identified in longitudinal plasma samples, additional mutations missed in tumor tissues were reported in 3/21 patients (14 %), while emerging mutations were reported in 9/21 patients (43 %). In the postoperative surveillance setting, ctDNA-based MRD detection anticipated clinical recurrence with a median lead-time of 9.9 months (interquartile range, 8.0–14.5 months) in 17/27 patients (63 %). When detected within 14 weeks after surgery, MRD correlated with disease recurrence after adjusting for classical prognostic variables (HR = 3.0; 95 % CI, 1.1–7.9; p = 0.03).
Conclusions
ctDNA detection is a useful biomarker for ITH and MRD in resectable HNSCC patients.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common cancer worldwide . Around 50 % of locally advanced HNSCC recur after primary treatment, at the locoregional level or distantly . Locoregional recurrence can still be treated with salvage surgery or re-irradiation, both of which being associated with a reduced mortality risk by 49 % and 28 % respectively . In contrast, patients who are not amenable to local treatments have a dismal prognosis (i.e., 6 to 9 months in the absence of treatment) . The current treatment strategy is based on tumor location and disease stage and does not take into account the underlying etiology of HNSCC (e.g., infection with high-risk human papillomavirus [HPV] or heavy use of tobacco and alcohol) , although HPV has been shown to be associated with a better prognosis .
HNSCC is characterized by substantial heterogeneity at the molecular level . Tumor heterogeneity has been associated with a dismal prognosis due to decreased response to treatment and higher rates of relapse, even in HPV-positive HNSCC patients . The natural course of the disease and treatment pressure may select specific tumor clones with molecular characteristics that may impact patients prognosis . Treatment response assessment relies on conventional imaging and positron-emission tomography, which cannot detect disease at the molecular level. In addition, the characterization of the mutational landscape for guiding precision medicine theoretically requires invasive and repeated tissue biopsies, which might not capture spatial intra-tumor heterogeneity or clonal dynamics over time.
A non-invasive way to overcome these limitations is the detection and analysis of circulating tumor DNA (ctDNA), a fraction of double-stranded DNA fragments that are freely released from tumor cells into the bloodstream . ctDNA detection methods often rely on identification of tumor-specific alterations using digital PCR or next-generation sequencing (NGS) techniques. Such targeted approaches have been successfully implemented in small proof of concept studies in HNSCC for minimal residual disease (MRD) detection following curative-intent treatment . On the other hand, tissue-agnostic methods (e.g., without prior knowledge of somatic mutations in the solid tumor) have enabled identification of potentially actionable mutations and emerging resistant tumor subclones in patients with advanced HNSCC .
We hypothesized that targeted NGS for ctDNA sequencing could be effective in detecting MRD while providing clinically relevant insights on tumor heterogeneity in non-metastatic resectable HNSCC. Based on the prospective SCANDARE HNSCC patients’ cohort (NCT03017573) treated with curative-intent upfront surgery, we sought to assess (i) the feasibility of a dedicated targeted NGS approach for ctDNA detection and monitoring, (ii) the potential of ctDNA as a biomarker of tumor heterogeneity, and (iii) the clinical validity of ctDNA-based MRD detection.
Patients and methods
Patients and sample collection
Tumor tissues and blood samples were obtained from HNSCC patients treated with upfront curative-intent surgery at Institut Curie (Paris, France) between January 17, 2017, and July 12, 2019, in the prospective biobanking SCANDARE study. Patients were chosen retrospectively from the biorepository to enrich for recurrences. SCANDARE was approved by a national ethics committee (CPP Ile-de-France 3) and by the National Agency for the Safety of Drugs and Health Products (ANSM).
All patients provided formalin-fixed paraffin-embedded (FFPE) blocks of the primary resected tumor specimen, altogether with ≥2 blood samples (including one blood sample collected at surgery). For each patient, one to five geographically separate region(s) of the tumor body were delineated by a pathologist (median [interquartile range, IQR] number, 4 [2–4] regions/patient), and further sequenced by using multiregion targeted NGS. The tumor region with the highest number of pathogenic variants was prioritized to infer the mutational landscape of the whole disease, and to design a dedicated targeted assay-panel for ctDNA sequencing. In additional analysis, data acquired from sequencing non-prioritized tumor regions was manually inspected to control for false-positive tumor heterogeneity assumptions.
Blood samples were collected in EDTA tubes and centrifugated within 3 h from sampling. Blood samples were collected at the time of surgical removal of the primary tumor, within 14 weeks after surgery (median [IQR] time, 3.4 [2.5–4.9] weeks), at six months and at recurrence, if any.
All patients received standard adjuvant therapy (e.g., radiotherapy with or without chemotherapy) or clinical follow-up according to the stage of the disease and usual procedures. A three-year follow-up period was conducted as part of the clinical routine.
Targeted NGS on tissue
Targeted NGS on tumor tissues was performed by using a custom panel of 571 genes (named DRAGON), covering a list of frequently mutated tumor suppressor genes (TSG) and oncogenes in HNSCC. Genes were classified regarding the literature and databases (cBioPortal and OncoKB ) in TSG, oncogenes, and genes considered as both an oncogene and a TSG. DNA sequencing protocol, data processing and bioinformatics analyses are summarized in the Supplementary Materials .
Targeted NGS on plasma
NGS-based ctDNA analysis was performed by using an amplicon-based targeted sequencing test (named OncoFOLLOW™). Somatic variants identified in tumor tissues by targeted NGS were prioritised to build a patient-specific panel, which included a median of 14 variants (IQR, 13–16 variants). This custom panel was combined with a fixed core panel of 40 genes, covering a list of frequently mutated tumor suppressor genes and oncogenes in HNSCC ( Supplementary Table 1 ). ctDNA sequencing protocol and variant allele frequency (VAF) estimation are summarized in the Supplementary Materials .
Statistical analyses
This report was written in accordance with REMARK criteria .
Endpoints included the sensitivity of the dedicated ctDNA assay-panel for ctDNA detection at surgery, in postoperative blood samples and at recurrence; the concordance rates of mutations between tumor tissues and ctDNA at surgery and at recurrence; the lead-time defined as the time interval (in months) between the first ctDNA-positive postoperative blood sample and the time of clinically or radiologically detected disease progression; recurrence-free survival (RFS) in MRD-positive and MRD-negative patients as assessed by using Kaplan-Meier methods and Cox proportional hazard regression models. MRD was defined by the presence of ctDNA in postoperative blood samples that typically preceded clinical relapse by several months. RFS was defined as the time elapsed between the date of inclusion and the date of cancer recurrence or last follow-up. Overall survival (OS) was defined as the time elapsed between the date of inclusion and death from any cause or last follow-up.
Results
Patient characteristics and sample collection
Forty-three HNSCC patients treated with upfront curative-intent surgery in the prospective biobanking SCANDARE study were included. Multiregion targeted NGS was performed on tumor tissues obtained at the primary surgery by using a custom panel of 571 genes. Two patients did not pass our filtering criteria due to low-coverage sequencing (1000x read depth <5 %) and aberrant tumor mutational burden (TMB) >400 Mut/Mb. Forty-one patients displayed trackable mutations on tumor tissues, which were further prioritised to build patient-specific panels of up to 21 variants combined with a fixed core panel of 40 genes for targeted NGS on plasma. Among them, 31 patients (31/41, 76 %) showed recurrent disease at a median follow-up of 41 months (IQR, 40–50 months, Figure 1 ).
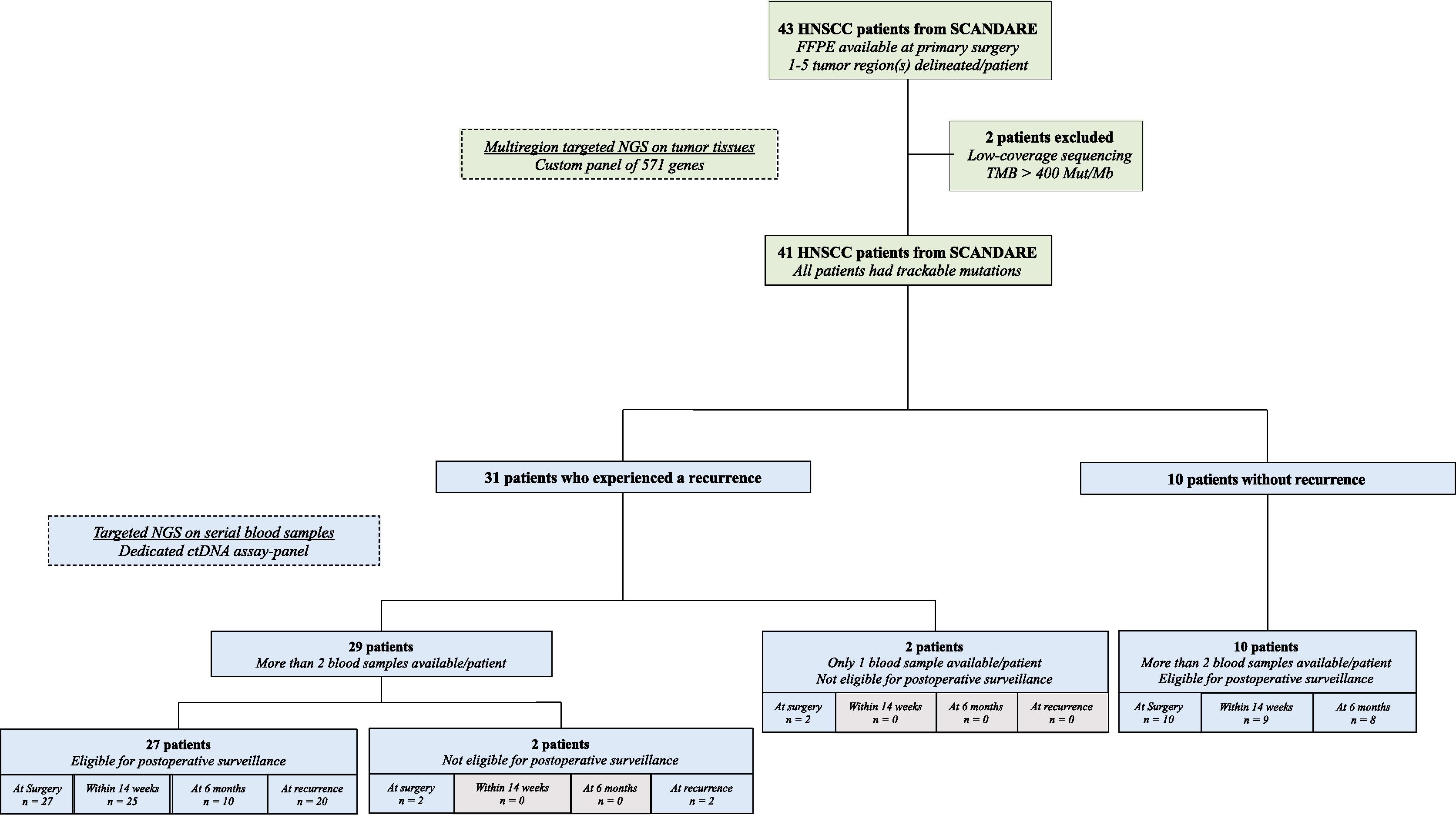
Serial plasma samples were collected at surgery ( n = 41, 100 %), within 14 weeks after surgery (34/41, 83 %), at six months (18/41, 44 %), and at recurrence (22/31, 71 %). In total, 41 patients at surgery and 22 patients at recurrence were analyzed for ctDNA profiling. Thirty-seven patients (37/41, 90 %) displayed plasma samples within 14 weeks after surgery and/or at six months and were monitored by targeted sequencing of ctDNA for postoperative surveillance ( Figure 1 ).
Baseline patient characteristics according to recurrence status are summarized in Table 1 .
| n (%) | Patients with a recurrence | Patients without recurrence | Total | p -value ¶ |
|---|---|---|---|---|
| ( n = 31) | ( n = 10) | ( n = 41) | ||
| Age, years | 1 | |||
| Median (IQR) | 66 (57–75) | 63 (58–79) | 65 (58–75) | |
| Sex, n (%) | 1 | |||
| Male | 25 (81 %) | 8 (80 %) | 33 (80 %) | |
| Female | 6 (19 %) | 2 (20 %) | 8 (20 %) | |
| Smoker, n (%) | 0.3 | |||
| Current/Former | 15 (48 %) | 7 (70 %) | 22 (54 %) | |
| Never | 16 (52 %) | 3 (30 %) | 19 (46 %) | |
| Alcohol consumption, n (%) | 1 | |||
| Current/Former | 15 (48 %) | 5 (50 %) | 20 (49 %) | |
| Never | 16 (52 %) | 5 (50 %) | 21 (51 %) | |
| Tumor location, n (%) | 0.07 | |||
| Oral cavity | 12 (39 %) | 8 (80 %) | 20 (49 %) | |
| Oropharynx | 15 (48 %) | 1 (10 %) | 16 (39 %) | |
| Larynx | 3 (10 %) | 0 | 3 (7 %) | |
| Hypopharynx | 1 (3 %) | 1 (10 %) | 2 (5 %) | |
| Grading, n (%) | 1 | |||
| Well/Moderately differentiated | 27 (87 %) | 9 (90 %) | 36 (88 %) | |
| Poorly/Undifferentiated | 4 (13 %) | 1 (10 %) | 5 (12 %) | |
| HPV status, n (%) | 1 | |||
| HPV negative | 23 (74 %) | 7 (70 %) | 30 (73 %) | |
| HPV positive | 8 (26 %) | 3 (30 %) | 11 (27 %) | |
| T classification, n (%) | 0.5 | |||
| T1/T2 | 16 (52 %) | 7 (70 %) | 23 (56 %) | |
| T3/T4 | 15 (48 %) | 3 (30 %) | 18 (44 %) | |
| Lymph node involvment, n (%) | 0.5 | |||
| Negative | 13 (42 %) | 6 (60 %) | 19 (46 %) | |
| Positive | 18 (58 %) | 4 (40 %) | 22 (54 %) | |
| Adjuvant therapy, n (%) | 0.6 | |||
| None | 11 (35 %) | 2 (20 %) | 13 (32 %) | |
| Radiotherapy alone | 13 (42 %) | 3 (30 %) | 16 (39 %) | |
| Chemoradiation | 7 (23 %) | 5 (50 %) | 12 (29 %) |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree