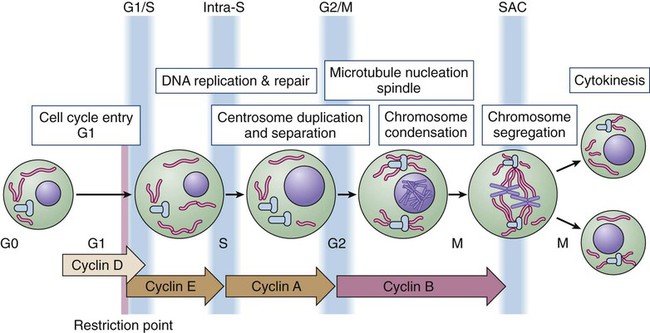
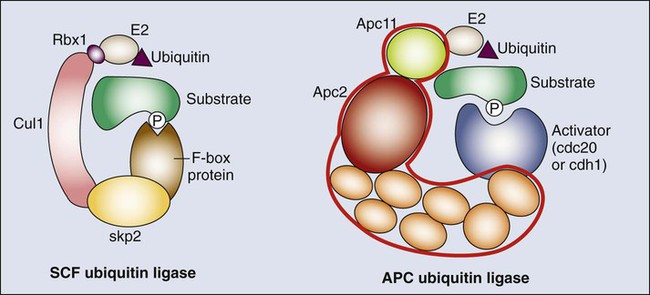
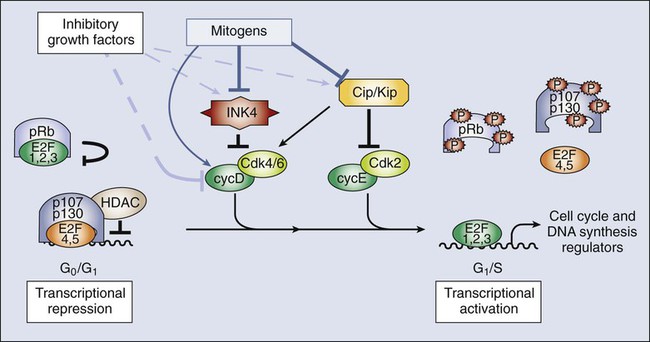
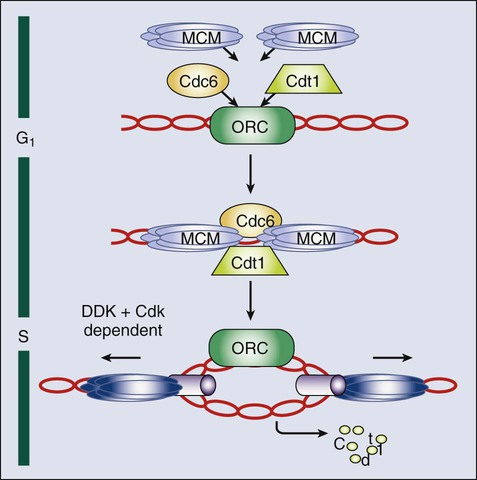
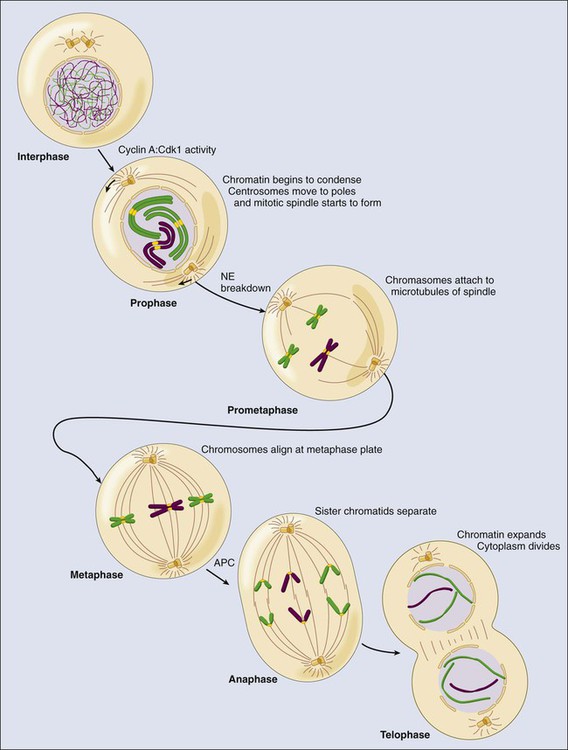
• Most cells in postnatal tissues are quiescent. Exceptions include abundant cells of the hematopoietic system, skin, and gastrointestinal mucosa, as well as other minor progenitor populations in other tissues. • Many quiescent cells can reenter into the cell cycle with the appropriate stimuli, and the control of this process is essential for tissue homeostasis. • The key challenges for proliferating cells are to make an accurate copy of the 3 billion bases of DNA (S phase) and to segregate the duplicated chromosomes equally into daughter cells (mitosis). • Progression through the cell cycle is dependent on both intrinsic and extrinsic factors, such as growth factor or cytokine exposure, cell-to-cell contact, and basement membrane attachments. • The internal cell cycle machinery is controlled largely by oscillating levels of cyclin proteins and by modulation of cyclin-dependent kinase (Cdk) activity. One way in which growth factors regulate cell cycle progression is by affecting the levels of the D-type cyclins, Cdk activity, and the function of the retinoblastoma protein. • Cell cycle checkpoints are surveillance mechanisms that link the rate of cell cycle transitions to the timely and accurate completion of prior dependent events. p53 is a checkpoint protein that induces cell cycle arrest, senescence, or death in response to cellular stress. • Checkpoints minimize replication and segregation of damaged DNA or the abnormal segregation of chromosomes to daughter cells, thus protecting cells against genome instability. • Disruption of cell cycle controls is a hallmark of all malignant cells. Frequent tumor-associated alterations include aberrations in growth factor signaling pathways, dysregulation of the core cell cycle machinery, and/or disruption of cell cycle checkpoint controls. • Because cell cycle control is disrupted in virtually all tumor types, the cell cycle machinery provides multiple therapeutic opportunities. Most cells in the adult body are quiescent—that is, they are biochemically and functionally active but do not divide to generate daughter cells. However, specific populations retain the ability to proliferate throughout the adult life span, which is essential for proper tissue homeostasis. For example, cells of the hematopoietic compartment and the gut have a high rate of turnover, and active proliferation is therefore essential for the maintenance of these tissues. On average, about 2 trillion cell divisions occur in an adult human every 24 hours (about 25 million per second). The decision about whether to proliferate is tightly regulated.1,2 It is influenced by a variety of exogenous signals, including nutrients and growth factors, as well inhibitory factors and the interaction of the cell with its neighbors and with the underlying extracellular matrix. Each of these factors stimulates intracellular signaling pathways that can either promote or suppress proliferation. The cell integrates all of these signals, and if the balance is favorable, the cell will initiate the proliferation process. During the past three decades, extensive effort has been placed on unraveling the basic molecular events that control this process. Studies in a variety of organisms have identified evolutionarily conserved machinery that controls eukaryotic cell cycle transitions through the action of key enzymes, including cyclin-dependent kinases (Cdks) and other kinases.3 It is essential that proliferating cells copy their genomes and segregate them to the daughter cells with high fidelity. Eukaryotic cells therefore have evolved a series of surveillance pathways, termed “cell cycle checkpoints,” that monitor for potential problems during the cell cycle process.4 Human cells are continuously exposed to external agents (e.g., reactive chemicals and ultraviolet light) and to internal agents (e.g., byproducts of normal intracellular metabolism, such as reactive oxygen intermediates) that can induce DNA damage. The cell cycle checkpoints detect DNA damage and activate cell cycle arrest and DNA repair mechanisms, thereby maintaining genomic integrity. Anything that disrupts proper cell cycle progression can lead to either the reduction or expansion of a particular cell population. It is now clear that such changes are a hallmark of tumor cells, which carry mutations that impair signaling pathways; that is, they suppress proliferation and/or activate pathways that promote proliferation. In addition, most (if not all) human tumor cells have mutations within key components of both the cell cycle machinery and checkpoint pathways.1,5,6 This characteristic has important clinical implications, because the presence of these defects can modulate cellular sensitivity to chemotherapeutic regimens that induce DNA damage or mitotic catastrophe. This chapter focuses on the mechanics of the cell cycle and checkpoint-signaling pathways and discusses how this knowledge can lead to the efficient use of current anticancer therapies and to the development of novel agents. Cell division proceeds through a well-defined series of stages (Fig. 4-1). First, the cell moves from the quiescent (also known as G0) state into the first gap phase, or G1, in which the cell essentially is readying itself for the cell division process. This process involves a dramatic upregulation of both transcriptional and translational programs not only to yield the proteins required to regulate cell division but also to essentially double the complement of macromolecules so that one cell can give rise to two cells without a loss of cell size. Not surprisingly, this process takes a significant amount of time (from 8 to 30 hours) and energy. Studies with cultured cells show that mitogenic growth factors are essential for continued passage through the G1 phase. Specifically, if growth factors are withdrawn at any point during this phase, the cell will not divide. However, as it nears the end of the G1 phase, the cell passes through a key transition point, called the restriction point, whereupon it becomes growth factor independent and is fully committed to undergoing cell division.1 Within an hour or two, the cell enters the synthesis phase, or S phase, in which each of the chromosomes is replicated once and only once. The cell then enters a second gap phase, called G2, which lasts 3 to 5 hours, and then initiates mitosis, or the M phase, a rapid phase (lasting about 1 hour) in which the chromosomes are segregated. Upon completion of mitosis, the daughter cells can enter quiescence or initiate a second round of cell division, depending on the milieu. Progression throughout the different phases of the cell cycle depends on the activity of key molecules that drive transcription, translation, or the structural changes required for cell division (Table 4-1). A large number of these changes are modulated by protein phosphorylation and dephosphorylation, but other molecular processes such as SUMOylation, acetylation, or ubiquitin-dependent protein degradation are crucial for ordered cell cycle progression. Many of these cell cycle regulators have been involved in tumor development or may be attractive targets for cancer therapy and will be introduced in the following sections. Table 4-1 Molecules Involved in Cell Cycle Regulation The Cdks constitute a large subfamily of highly conserved Ser/Thr kinases that are defined by their dependence on a regulatory subunit, called a cyclin.9–9 The first identified human Cdk, called Cdk1 (originally cdc2), was cloned by virtue of its ability to complement a mutant cdc2 yeast strain.10 Subsequent studies identified additional human Cdks and determined that they regulate distinct cell cycle stages; for example, Cdk4 and Cdk6 regulate cell cycle entry, whereas Cdk2 may have specific roles during the G1-to-S transition and S phase. Cdk1 is essential in the control of G2 and mitosis and also may play additional roles in earlier stages. The human genome encodes about 15 additional Cdks, although the functional relevance of many of them is still unknown.3,8,9 The activity of these kinases is controlled by multiple regulatory mechanisms. Cdks act in association with a cyclin subunit that binds to the conserved PSTAIRE helix within the kinase.11 Cyclin binding causes a reorientation of residues within the active sites that is essential for kinase activity.7,11 The associated cyclin also determines the substrate specificity of the resulting cyclin-Cdk complex. The cyclins are quite divergent, especially in their N-terminal sequences, but they all share a highly conserved 100-amino acid sequence, called the cyclin box, that mediates Cdk binding and activation.8 As their name implies, cyclins originally were identified as proteins whose expression was restricted to a particular stage of the cell cycle12 because of cell cycle–dependent regulation of both cyclin gene transcription and protein degradation. The human genome encodes more than 25 cyclin-like proteins, yet only four distinct subclasses—D-, E-, A-, and B-type cyclins—are involved in cell cycle regulation (see Fig. 4-1).8 Each of these classes has a few paralogs (e.g., cyclin D1, D2, and D3; cyclin E1 and E2; cyclin A1 and A2; and cyclin B1, B2, and B3). The relative roles of these paralogs are not completely clear in most cases. Although some functional redundancy may exist, published evidence suggests differences in regulation, expression pattern, and substrate specificity.8,13 The activation of cyclin-Cdk complexes requires considerable posttranslational regulation.8,14 First, kinase activation is dependent on phosphorylation of a threonine residue that is adjacent to the active site (Thr160 in Cdk2). This phosphorylation is catalyzed by a kinase, called Cdk-activating kinase (CAK).15,16 In mammalian cells, phosphorylation occurs after cyclin binding. Although it appears that at least two mammalian CAKs exist, the major CAK is a trimolecular complex composed of Cdk7, cyclin H, and Mat1. The Cdk7-cyclinH-Mat1 complex also is required for the control of basal transcription via regulation of RNA polymerase II function.16 Second, when it is first formed, the cyclin-Cdk complex frequently is subject to inhibitory phosphorylation of Thr14 and Tyr15 residues within the Cdk’s active site by the Wee1 (Tyr15) and Myt1 (Thr14 and Tyr15) kinases.8,17 Activation of the cyclin/Cdk complex is then dependent on the action of a dual-specificity phosphatase called Cdc25. Mammalian cells have three different Cdc25 proteins, called Cdc25a, Cdc25b, and Cdc25c, which show some specificity for different cyclin-Cdk complexes.18 Cdks are modulated by a series of CdK inhibitors (CKIs) that play a key role in establishing the activity of the cyclin-Cdk complexes in response to either external signals or internal stresses.19 The CKIs can be divided into two distinct families based on their biological properties. The first CKI family is named INK4, based on their roles as INhibitors of CDK4. The INK4 family has four members called p16INK4a, p15INK4b, p18INK4c, and p19INK4d (encoded by the CDKN2A-D genes in humans). These INK4 proteins specifically prevent the binding of cyclins to monomeric Cdk4 and Cdk6 but do not inhibit other Cdks.11,19 The second CKI family is named Cip/Kip and includes three members: p21Cip1 (also called p21Waf1), p27Kip1, and p57Kip2 (encoded by the CDKN1A-C genes in humans).19 These Cip/Kip proteins have two major activities. First, they do not bind to monomeric Cdks but associate with and inhibit the activity of cyclin-Cdk complexes already formed. Second, Cip/Kip proteins may promote the assembly of cyclin D-Cdk4/6 complexes without dramatically perturbing its kinase activity.19,20 This activity is somehow modulated by phosphorylation of Cip/Kip proteins by Src, Jak2, and Akt kinases,21–24 directly linking Cdk regulation with the activity of these upstream mitogenic pathways. In addition to regulating the cell cycle, Cip/Kip proteins play important roles in apoptosis, transcriptional regulation, cell fate determination, cell migration, and cytoskeletal dynamics.25,26 The retinoblastoma protein (pRb) originally was identified by virtue of its association with hereditary retinoblastoma.27 It behaves as a classic tumor suppressor: Affected persons inherit a germline mutation within one RB1 allele, and loss of heterozygosity is seen in all of the tumors. Subsequent studies showed that the transforming ability of small DNA tumor viruses, including human papilloma virus, adenovirus, and simian virus, was dependent on the ability of virally encoded oncoproteins (E7, E1A, and SV40, respectively) to bind and inhibit pRb.28,29 Moreover, the RB1 gene is inactivated in approximately one third of all sporadic human tumors. pRb and the pRb-related proteins p107 and p130, collectively known as the pocket proteins, are transcriptional repressors whose major function is to inhibit the expression of cell-cycle related proteins (Table 4-1).30 This suppressive activity is largely dependent on the ability to prevent cell cycle entry through inhibition of the E2F transcription factors.30,31 The E2F proteins regulate the cell cycle–dependent transcription of numerous targets, including core components of the cell cycle control (e.g., cyclin E and cyclin A) and DNA replication (e.g., Cdc6, Cdt1, and the Mcm proteins) machineries.31,32 pRb regulates E2F through two distinct mechanisms. First, its association with E2F is sufficient to block the transcriptional activity of E2F.33,34 Second, the pRb-E2F complex can recruit histone deacetylases to the promoters of E2F-responsive genes and thereby actively repress their transcription.35,36 Cell cycle entry requires the phosphorylation of pRb by cyclin-Cdk complexes and the consequent dissociation of pRb from E2F.1 Studies to date have identified eight E2F genes that encode nine different E2F proteins.31,32 Pocket proteins can regulate a subset of these factors: E2F1, E2F2, E2F3a, E2F3b, E2F4, and E2F5. These E2F proteins associate with a dimerization partner, called DP, and the resulting complexes function primarily as either activators (E2F1, E2F2, and E2F3a) or repressors (E2F4 and E2F5) of transcription under the direction of the pocket proteins. Recent observations suggest that several of these factors may act either as positive or negative regulators of transcription depending on the cell type or the differentiation state.31,37–39 Most classic E2F target genes are regulated by the coordinated action of these repressor and activator E2Fs. Cdc14 phosphatases are the major Cdk-counteracting proteins in yeast. Although two family members exist in mammals, their relevance in the cell cycle is not well understood.40,41 In eukaryotes, two major complexes, PP1 and PP2A, account for more than 90% of protein phosphatase activity. In fact, these enzymes correspond to hundreds of phosphatase complexes assembled from a few catalytic subunits (PP1α, PP1β/δ, and PP1γ1/2 for PP1, and the Cα and Cβ isoforms for PP2A) and a diverse array of regulatory subunits.42 Recent evidence suggests that these protein families cooperate in the dephosphorylation of most cell cycle kinase targets, including the retinoblastoma family or mitotic phosphoproteins.40,43,44 PP1 and PP2A are major phosphatases responsible for pRb dephosphorylation during mitotic exit, although the relative roles of these complexes or the particular holoenzymes involved are not clear.43,44 Similarly, both PP1 and PP2A are required for dephosphorylation of hundreds of mitotic proteins that are phosphorylated by Cdk1, as well as the other mitotic kinases. Thus it has been suggested that the cell cycle ultimately is regulated by the dynamic equilibrium between Cdks (and partially by the other mitotic kinases) and PP1/PP2A activity. In the absence of Cdk activity, the balance tilts in favor of the phosphatases. When Cdks are activated, phosphatase activity is overtaken. Cdk1 is able to directly inhibit PP1 by direct phosphorylation of the catalytic subunit. The Cdk-dependent inhibition of PP2A, on the other hand, is not direct; rather, it is mediated by a new kinase known as Greatwall in flies and Xenopus or Mastl in mammals.47–47 Cdk1 phosphorylates and activates Mastl, which in turn phosphorylates Arpp-19 and Ensa, two highly related proteins that function as inhibitors of a particular PP2A holoenzyme encompassing a regulatory subunit of the B55 family.45,46,48,49 Reactivation of PP1 and PP2A phosphatases is a mandatory step for the exit from mitosis and the transition to interphase.52–52 The original observation that cyclin levels are tightly regulated during the cell cycle implies that these proteins are not only regulated at the transcriptional level but also at the protein level. It is now evident that ubiquitin-mediated protein degradation is a major regulatory mechanism to ensure ordered transition through the different phases of the cell division cycle. Ubiquitylation depends on an enzymatic cascade, in which ubiquitin ligases recruit specific substrates for modification. About 600 ubiquitin ligases are encoded by the human genome. Among them, the Skp1–Cullin1–F-box (SCF) and the anaphase-promoting complex/cyclosome (APC/C) are known for driving the degradation of cell cycle regulators to accomplish irreversible cell cycle transitions.55–55 SCF has three core components: a RING finger protein, called Rbx1, which recruits the E2-ubiquitin conjugate; a cullin (Cul1); and Skp1 (Fig. 4-2).54 Skp1 acts to recruit a family of proteins, called F-box proteins, that determines the target specificity of the SCF complex. Once SCF binds its substrate, it transfers a ubiquitin molecule to lysine residues within the target protein to create a polyubiquitin chain, which targets the substrate to the proteasome for degradation.56 The APC/C is a much larger complex, but it also contains a RING finger protein, called Apc11, to recruit the E2-ubiquitin conjugate, and a core cullin subunit (Apc2). In addition, APC/C is activated by a cofactor that, in a manner comparable with that of the F-box proteins of SCF, establishes substrate specificity (see Fig. 4-2).57 Cdc20 is the mitotic cofactor of APC/C, and it targets several cell cycle regulators (A- and B-type cyclins, Nek2, and securin) during mitotic entry and the metaphase-to-anaphase transition. Cdh1 (also known as FZR1 in mammals) replaces Cdc20 during mitotic exit and is the cofactor responsible for the elimination of many cell cycle regulators in the G0 or G1 phase to prevent unscheduled DNA replication.57,58 Recent studies have provided new insights into the intricate relationship between ubiquitylation and the cell division apparatus, including new roles for atypical ubiquitin chains, new mechanisms of regulation, and extensive cross talk between ubiquitylation enzymes.56 In addition to the central role of Cdks in the cell cycle, many other kinases play critical roles in cell cycle progression.3 Aurora and Polo kinases were first identified in genetic studies in flies as a result of their essential role in mitotic progression.61–61 Each of these families of kinases is represented by a single member in yeast, whereas three Aurora kinases (Aurora-A, Aurora-B, and Aurora-C) and 5 Polo-like kinases (Plk1 through Plk5) exist in humans (Table 4-1).60,62 Aurora A participates in several processes required for building a bipolar spindle, including centrosome separation and microtubule dynamics. Aurora A is activated by Tpx2, and the Aurora A–Tpx2 holoenzyme may have critical implications in tumor development.63 Aurora B, on the other hand, is part of the chromosome passenger complex (CPC) that localizes to the kinetochores from prophase to metaphase and to the central spindle and midbody in cytokinesis. Other components of the CPC include Incenp, survivin, and borealin, and this complex regulates proper microtubule-kinetochore attachment and promotes biorientation during mitosis.61 Aurora C may play similar roles to Aurora B, although it is mostly expressed in germ cells and may play a specific role in meiosis and during the first embryonic cycles.66–66 Plk1 functions as a major regulator of centrosome maturation, mitotic entry, and cytokinesis, whereas Plk4 is a critical regulator of centriole duplication.59,67–69 The other members of the family, Plk2, Plk3, and Plk5, are mostly involved in stress responses during interphase or in neuron biology.3,62 A different group of additional cell cycle kinases with potential interest in tumor biology is the never in mitosis, gene A (NIMA)-related kinase family (Nek1 through Nek11). Four of these proteins, Nek2, Nek6, Nek7, and Nek9, are involved in cell cycle progression, whereas all other family members are likely to play critical roles in cilia and centrioles.3,70,71 Nek2, the closest relative to the Aspergillus NIMA kinase, localizes to the centrosome and plays a role in establishing the bipolar spindle by initiating the separation of centrosomes and contributing to microtubule organization at the G2/M transition by phosphorylating several centrosomal substrates. Nek2 also may play additional roles in chromosome condensation and the mitotic checkpoint. Nek9, Nek6, and Nek7 function in a kinase cascade that participates in centrosome separation and the formation and/or maintenance of the mitotic spindle.70,72 The activity of all these kinases is functionally linked to many other microtubule-associated proteins such as kinesin motor proteins, which determine the dynamic behavior of the mitotic spindle required for chromosome movement during mitosis.73,74 Because most adult cells are quiescent, the mechanisms that determine their quiescent state and their reentry into the cell cycle upon the appropriate stimuli are key determinants of tissue homeostasis. In quiescent cells, several DP-E2F complexes associate with the promoters of E2F-responsive genes and recruit pocket proteins, along with their associated histone deacetylases, to actively repress their transcription.75,76 This repression machinery therefore prevents the expression of proteins required for DNA synthesis and chromosome segregation. In addition, CKIs normally are expressed in quiescent cells, preventing the activation of Cdks.19 D-type cyclins are present at very low levels in most quiescent cells, in large part because they are phosphorylated by an abundant kinase called Gsk3β and then exported to the cytoplasm for degradation.77 In response to mitogenic signaling, the activity of Gsk3β is inhibited, preventing the degradation of D-type cyclins. In addition, mitogens directly induce the transcription of this class of cyclins. Notably, individual mitogenic signaling pathways not only cooperate in inducing a single D-type cyclin but also may induce different D-type cyclins.78,79 Transcriptional induction of D-type cyclins promotes cell cycle entry through two distinct mechanisms. First, D-type cyclins associate with Cdk4 and Cdk6, and the resulting complexes phosphorylate pRb, partially inactivating its transcriptional suppressor function (Fig. 4-3).80–83 Second, the D-type cyclins titrate CKIs away from other Cdks and thereby promote their activation.19,84 Cdk activity increases, and the phosphorylation of the pocket proteins causes them to release their associated DP-E2Fs. Repressive E2Fs such as E2F4 and E2F5 dissociate from the DNA, and the free E2F complexes—DP-E2F1, DP-E2F2, and DP-E2F3—now occupy the promoters and activate their transcription. Cyclin E is itself an E2F-responsive gene, and this regulation creates a strong feed-forward loop. Cyclin E binds to Cdk2, and this complex may promote further pRb inactivation.85,86 Cyclin E–Cdk2 also phosphorylates the Cdk inhibitor p27Kip1 on Thr187.87,88 This action creates a high-affinity binding site for the SCF ubiquitin ligase bound to the F-box protein Skp2, leading to p27Kip1 degradation during the G1/S transition.89–92 Finally, cyclin E-Cdk2 phosphorylates itself on multiple sites, creating a recognition site for SCF-Fbw7/Cdc4 and thereby ensuring its own destruction.93,94 The fact that lack of Cdk2 in the mouse does not result in defective mitotic cycles suggests that the activity of this protein overlaps with other Cdks, with Cdk1 being the best candidate.97–97 Indeed, Cdk1 is able to bind interphase cyclins such as cyclin D and cyclin E, and it is sufficient for G1/S transition, at least in the absence of other interphase Cdks.98,99 On the other hand, cyclin E–Cdk2 may have specific functions in the duplication of centrosomes required for formation of the mitotic spindle,102–102 and it has an essential role in meiosis, although the molecular basis for this requirement is not fully understood.96,103–105 The DNA replication machinery is optimized to ensure that the genome is copied once—and only once—in each cell cycle.106 This optimization is achieved through a two-step process that first establishes a prereplication complex (pre-RC) at each origin of replication, a process that is frequently referred to as origin licensing, and subsequently transforms pre-RCs into the preinitiation complex (pre-IC) that activates DNA replication (Fig. 4-4). These two steps occur at distinct stages of the cell cycle to ensure that origins are only licensed once per cell cycle and rereplication cannot occur. Pre-RC formation takes place during G1. The first step in this process is the recruitment of the multiprotein complex called the origin recognition complex to the origin DNA.107 The origin recognition complex recruits additional proteins including Cdc6, Cdt1, and finally the mini chromosome maintenance (MCM) complex, a helicase that is required to unwind the DNA strands to form the pre-RC. Once cells enter S phase, the transformation of the pre-RC to the pre-IC requires the activity of two kinases: a Cdk and the Ddf4-dependent kinase, which is composed of the Dbf4 regulatory subunit and the Cdc7 kinase.108,109 The action of these kinases allows numerous additional proteins to associate with the pre-RC and form the pre-IC.110 Assembly of the pre-IC is thought to trigger DNA unwinding by the MCM complex, recruitment of the DNA polymerases, and initiation of the replication process, frequently called “origin firing.” The transformation of the pre-RC to the pre-IC can occur at different time points in S phase, depending on whether the origin fires early or late.106 The system can tolerate this heterogeneity because the pre-RC is disassembled after firing and cannot re-form until the subsequent cell cycle. This process occurs through several mechanisms. The MCM complex travels with the replication fork in its role as the DNA helicase. Some evidence also indicates that phosphorylation of Orc1 reduces its ability to bind to origins. Finally, and most important, Cdt1 is prevented from participating in pre-RC formation outside of the G1 phase in two distinct ways. First, Cdt1 is marked for destruction by ubiquitination.111 This process is mediated by SCF-Skp2 and particularly by an E4 ubiquitin ligase that includes Rbx1 (to recruit the E2-ubiquitin), a cullin (Cul4), Ddb1, and Dtl/Cdt2 (the substrate specificity factor).114–114 Importantly, this Cul4-Ddb1-Dtl/Cdt2 complex functions independently of Cdt1 phosphorylation. Instead, Cdt1 is targeted only when proliferative cell nuclear antigen is present on the DNA, which occurs primarily as a consequence of the initiation of DNA replication.115 Second, cells possess a protein called geminin that sequesters Cdt1 and prevents it from participating in pre-RC formation. Geminin is present specifically in S, G2, and early M phase cells. However, the APC/C ubiquitinates geminin and thereby triggers its destruction. This action creates a window between anaphase of mitosis and the late G1 phase (when APC/C-Cdh1 is inactivated) in which geminin is absent and therefore Cdt1 is free to participate in pre-RC formation.116 The A-type cyclins are first transcribed late during the G1 phase under the control of the E2F transcription factors in a similar manner to that of cyclin E. However, in contrast to cyclin E, cyclin A associates with both Cdk2 and Cdk1 and acts at two distinct cell cycle stages.117,118 First, cyclin A is required for S phase progression, which was established by showing that injection of either cyclin A antisense constructs or antibodies was sufficient to block S phase progression.117,119 Notably, cyclin A–Cdk2 enters the nucleus at the start of S phase and is specifically localized at nuclear replication foci, and thus it is thought to be actively involved in the firing of replication origins.120 As was described previously, cyclin A–Cdk2 also is required to phosphorylate E2F-1 and mediate its degradation, which is required to prevent E2F1 from triggering apoptosis.121,122 There are two A-type cyclins and three B-type cyclins with different expression patterns. Cyclin A1 and cyclin B2 are mostly expressed in meiotic cells, although these proteins also may be present in small quantities in mitotic cells.8 The analysis of mutant mouse models shows that cyclin A1 or B2 loss has no detectable effect on development, whereas cyclin A2 and B1 are absolutely required for embryogenesis.8 Cyclin B3 is an early meiotic cyclin that is expressed in the leptotene and zygotene phases of spermatogenesis.123,124 Interestingly, a new oncogenic fusion has been observed between BCOR (encoding the BCL6 co-repressor) and CCNB3 (encoding the testis-specific cyclin B3) on the X chromosome, indicating a biologically distinct class of sarcomas.125 Most of the information discussed in this chapter therefore relates to cyclin A2 and B1. Cyclin A2 is active during the G2 phase and at the beginning of mitosis.117 Here it is thought to play a key role in initiating the condensation of chromatin and also might participate in the activation of the cyclin B–Cdk1 complexes. Cyclin A2 is destroyed upon nuclear envelop breakdown in an APC/C-Cdc20–dependent manner.126 A-type cyclins are then substituted by B-type cyclins. Cyclin B1 protein accumulates steadily through the G2 phase and associates specifically with Cdk1, although it also may associate with Cdk2.118 However, the resulting cyclin B1–Cdk1 complex is mostly sequestered in the cytoplasm, and it is retained in an inactive form throughout the G2 phase via the inhibitory phosphorylation of Thr14 and Tyr15 in Cdk1’s active site by the Myt1 and, to a lesser extent, Wee1 kinases.129–129 Activation of cyclin B1–Cdk1 occurs in a highly synchronous manner during G2-prophase transition (see Fig. 4-1).130 This activation is mediated by two changes. First, the activities of Myt1 and Wee1 are downregulated at the transition between the G2 and M phases. Second, a dramatic increase in the activity of the Cdc25 phosphatases occurs that relieves the inhibitory phosphorylation of Thr14 and Tyr15. These activity changes are triggered by the phosphorylation of Myt1, Wee1, Cdc25a, and Cdc25c. Three different kinases are thought to contribute to this phosphorylation: Polo-like kinase (Plk1), cyclin A–Cdk1, and cyclin B1–Cdk1 itself. The involvement of cyclin B1–Cdk1 creates a powerful feed-forward loop; once a small amount of cyclin B1–Cdk1 is activated, it simultaneously inactivates its own inhibitors and activates its activators, enabling a rapid transformation of the entire cyclin B1–Cdk1 pool from the inactive state to the active state. Once active, cyclin B1–Cdk1 phosphorylates components of the centrosomes and initiates a process called centrosome separation, in which the centrosomes move to opposing poles of the nascent spindle, an event essential for formation of the mitotic spindle.131 Cyclin B1–Cdk1 then translocates across the nuclear membrane (which is still intact at this point in mitosis) to orchestrate mitotic events. Cdk1 phosphorylates nearly a hundred proteins and participates in such activities as chromosome condensation, nuclear envelop breakdown, modifications in the Golgi apparatus, and formation and dynamics of the mitotic spindle.8 Cdk1-deficient mouse embryos arrest at the two-cell state, indicating that this kinase is absolutely required for mitosis and cannot be compensated by other mammalian Cdks.99 The mitotic machinery is optimized to ensure that the replicated chromosomes are faithfully segregated to the daughter cells. This segregation is achieved through the use of a specialized microtubule-based structure, the mitotic spindle, on which the original chromosomes and their newly replicated copies, called sister chromatids, align and then are partitioned to opposite poles of the cell. The mitotic spindle is a highly dynamic structure that is maintained by many protein families, including motor molecules and other microtubule-associated proteins.132,133 The appropriate side-by-side alignment of the sister chromatids, termed biorientation, is facilitated by the physical tethering of the sister chromatids to one another. This process, called cohesion, actually occurs in S phase in a manner that is coordinated with the replication process.136–136 Cohesin is mediated by four proteins that together make up the cohesin complex. Two of these proteins, Smc1 and Smc3, have a long coiled-coil structure with a dimerization domain at one end that allows them to heterodimerize to form a V-like structure. Importantly, the remaining ends of Smc1 and Smc3 can associate with each another to form a functional adenosine triphosphate (ATP) domain. This domain acts in an ATP-dependent manner to recruit two additional proteins, Scc1 and Scc3, which form a closed-ring structure that most likely encircles the chromosomes.137,138 Cohesin loading onto chromosomes, catalyzed by a separate complex called kollerin, is thought to be mediated by the entry of DNA into cohesin rings, whereas dissociation, catalyzed by Wapl and several other cohesin subunits, is mediated by the subsequent exit of DNA.139 Largely on the basis of morphologic features, mitosis is divided into five different stages—prophase, prometaphase, metaphase, anaphase, and telophase (Fig. 4-5)—before separation of the daughter cells or cytokinesis. Prophase is essentially a preparative stage. One of the major events is modification of the DNA. In a process called resolution, the sister chromatids are untangled via the action of topoisomerase II.137,138 Resolution requires removal of the chromosome arm cohesin through phosphorylation of Scc3 by Plk1 and histone H3 by the aurora B kinase. Importantly, the cohesin complex at the centromere is somehow protected from this modification by a protein called shugosin (Sgo).142–142 In addition to resolution, the sisters undergo condensation—essentially, packaging into a more compact chromatin structure. This process involves two multimeric complexes, condensin I and II, which also contribute to sister chromatid resolution and requires phosphorylation by mitotic Cdks.137 During prophase, the nuclear envelope is still intact; consequently, differences in subcellular localization of the condensin and Cdk complexes allow only condensin II and cyclin A-Cdk1 (nuclear), and not condensin I and cyclin B-Cdk1 (cytoplasmic), to initiate condensation. The second major event in prophase is the translocation of the cytoplasmic cyclin B1-Cdk1 to the nucleus, where it phosphorylates components of the nuclear envelope and triggers its breakdown,143 which defines the transition from prophase to prometaphase. During prometaphase, the condensation process is accelerated because condensin I and cyclin B-Cdk1 now have access to the DNA. The sister chromatids become attached to spindle microtubules through a structure called the kinetochore, which is assembled onto centromeric DNA.146–146 Microtubules nucleated from the centrosomes attach to the kinetochore through a process called search and capture, in which individual microtubules grow and shrink until they contact and bind the kinteochore.145,147 Typically, one sister chromatid of the pair attaches first, and this attachment is further stabilized through the recruitment of additional microtubules from the same pole of the mitotic spindle to create a kinetochore fiber—that is, highly bundled microtubules bound to the kinetochore. The sister chromatids oscillate in the cell until the second sister chromatid is captured by microtubules emanating from the other pole. These oscillations continue until all of the chromosomes are properly aligned on the metaphase plate. Metaphase is defined as the point at which all of the chromosome pairs are fully condensed, attached to the mitotic spindle, and aligned at the center—termed the “metaphase plate.” The pulling of the kinetochore fibers toward the poles creates tension through the cohesin complex at the kinetochores, which indicates that the sister chromatids have achieved appropriate biorientation. The cell constantly monitors the attachments of microtubules to the chromosomes, and the tension that is generated by microtubules on the kinetochores ensures that the sister chromatids are properly aligned at the metaphase plate.148,149 This process is the basis of one of several cell cycle checkpoints, called the mitotic spindle assembly checkpoint (SAC), that will be described in more detail in the following sections. Anaphase is characterized by the segregation of the chromosomes.150 This event is controlled by the mitotic ubiquitin ligase APC/C-Cdc20. APC/C-Cdc20 ubiquitinates, and thereby triggers the degradation of, cyclin B1 and a protein called securin.53 Both securin and cyclin B1–Cdk1 complexes are able to bind and inhibit a protease called separase.53,150,151 APC/C-Cdc20 activity results in the degradation of cyclin B and securin and the subsequent separase activation. Once released, separase cleaves the Scc1 component of the cohesin complex, which opens the cohesin ring, unlinking the sister chromatids and allowing them to be pulled to opposite poles (see Fig. 4-5). The spindle poles then move farther apart to ensure that the chromosomes are fully segregated. The separase-dependent cleavage of Scc1 also is essential to link segregation of chromatids with the separation of centrioles during mitotic exit.152 During telophase, the mitotic spindle disassembles, leaving a single centrosome and a single set of chromosomes with each nascent daughter cell. Cyclin B1 degradation and the subsequent inactivation of Cdk1 results in DNA decondensation, and the nuclear envelope reforms around the segregated chromosomes to create two nuclei. These events are not only dependent on the loss of Cdk kinase activity, but the activation of mitotic exit phosphatases such as PP1 and PP2A is essential for the dephosphorylation of Cdk substrates.52–52 During anaphase, Cdh1, which is inhibited by Cdk-dependent phosphorylation during mitosis, is dephosphorylated and replaces Cdc20 as the main APC/C activator.53 APC/C-Cdh1 is responsible for the degradation of multiple cell cycle regulators, including Cdc20.53,58 APC/C-Cdh1 also activates the ubiquitination and degradation of geminin, allowing accumulation of Cdt1 for origin relicensing in the subsequent G1 phase, and the mitotic cyclins, allowing loss of Cdk kinase activity. Loss of Cdh1 does not result in major abnormalities during mitotic exit but results in earlier entry into the following S phase because of increased Cdk activity and DNA damage.153,154 Cdh1 therefore is required to prevent unscheduled entry into S phase and genomic instability.58 Finally, the cell undergoes cytokinesis, or cytoplasmic division. This process involves formation of a structure containing actin and myosin, called the contractile ring, on the inner face of the cell membrane. The position of the contractile ring is carefully controlled. For most mammalian cells, the ring begins to form in anaphase, and its position is established by the position of the metaphase plate. As the membrane grows, the contractile ring contracts steadily to form a constriction, termed the “cleavage furrow,” which ultimately separates the two nuclei and forms the two daughter cells.132,155,156 This process partially depends on several mitotic kinases such as Aurora B, a component of the CPC, and Plk1, which are located at the midbody. Plk1 controls the activity of RhoA guanosine triphosphatases, which are major regulators of the actomyosin ring required for abscission.67,157 Aurora B also ensures that abscission does not occur in the presence of DNA bridges to avoid DNA damage in a process known as the abscission checkpoint.156,158 At key transitions during eukaryotic cell cycle progression, signaling pathways monitor the successful completion of events in one phase of the cell cycle before proceeding to the next phase. These regulatory pathways are commonly referred to as cell cycle checkpoints.4 In a broader context, cell cycle checkpoints are signal transduction pathways that link the rate of cell cycle phase transitions to the timely and accurate completion of prior dependent events. Checkpoint surveillance functions are not confined to monitoring normal cell cycle progression; they also are activated by both external and internal stress signals. To minimize the possibility of errors, checkpoints exist at the four different phases of the cell cycle: G1 (to prevent entry into S phase), intra S phase, G2 (to prevent mitosis), and M (to avoid mitotic exit with abnormal segregation of chromosomes) (see Fig. 4-1).
Control of the Cell Cycle
Introduction
The Cell Division Cycle
Overview of the Cell Cycle Machinery
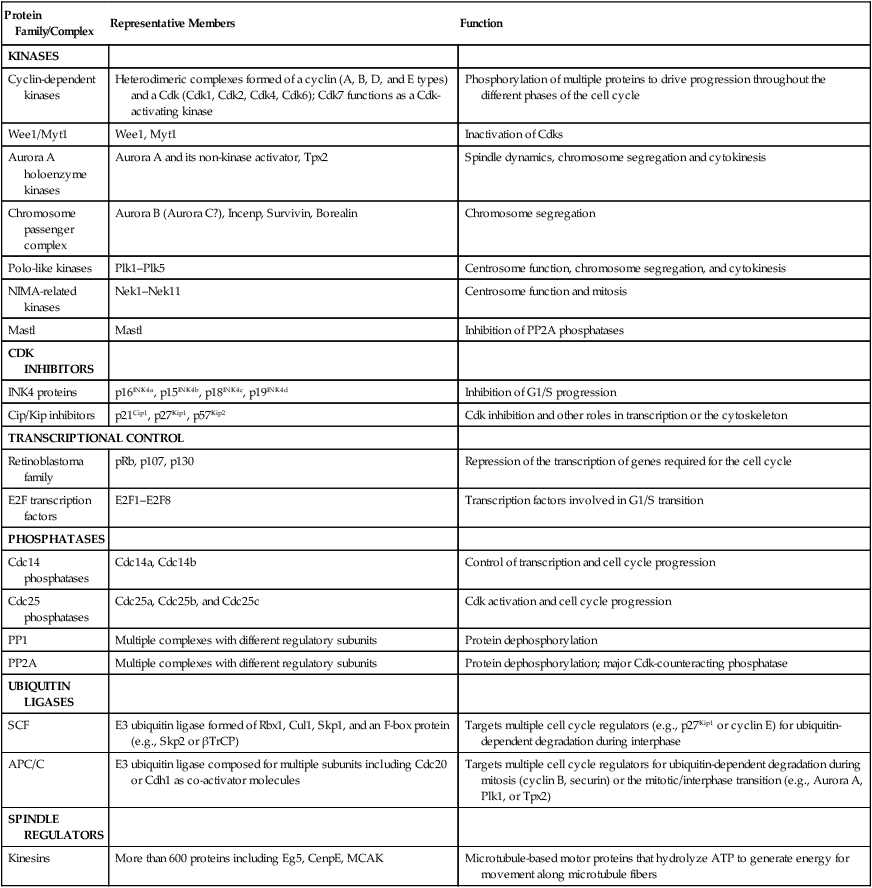
Protein Family/Complex
Representative Members
Function
KINASES
Cyclin-dependent kinases
Heterodimeric complexes formed of a cyclin (A, B, D, and E types) and a Cdk (Cdk1, Cdk2, Cdk4, Cdk6); Cdk7 functions as a Cdk-activating kinase
Phosphorylation of multiple proteins to drive progression throughout the different phases of the cell cycle
Wee1/Myt1
Wee1, Myt1
Inactivation of Cdks
Aurora A holoenzyme kinases
Aurora A and its non-kinase activator, Tpx2
Spindle dynamics, chromosome segregation and cytokinesis
Chromosome passenger complex
Aurora B (Aurora C?), Incenp, Survivin, Borealin
Chromosome segregation
Polo-like kinases
Plk1–Plk5
Centrosome function, chromosome segregation, and cytokinesis
NIMA-related kinases
Nek1–Nek11
Centrosome function and mitosis
Mastl
Mastl
Inhibition of PP2A phosphatases
CDK INHIBITORS
INK4 proteins
p16INK4a, p15INK4b, p18INK4c, p19INK4d
Inhibition of G1/S progression
Cip/Kip inhibitors
p21Cip1, p27Kip1, p57Kip2
Cdk inhibition and other roles in transcription or the cytoskeleton
TRANSCRIPTIONAL CONTROL
Retinoblastoma family
pRb, p107, p130
Repression of the transcription of genes required for the cell cycle
E2F transcription factors
E2F1–E2F8
Transcription factors involved in G1/S transition
PHOSPHATASES
Cdc14 phosphatases
Cdc14a, Cdc14b
Control of transcription and cell cycle progression
Cdc25 phosphatases
Cdc25a, Cdc25b, and Cdc25c
Cdk activation and cell cycle progression
PP1
Multiple complexes with different regulatory subunits
Protein dephosphorylation
PP2A
Multiple complexes with different regulatory subunits
Protein dephosphorylation; major Cdk-counteracting phosphatase
UBIQUITIN LIGASES
SCF
E3 ubiquitin ligase formed of Rbx1, Cul1, Skp1, and an F-box protein (e.g., Skp2 or βTrCP)
Targets multiple cell cycle regulators (e.g., p27Kip1 or cyclin E) for ubiquitin-dependent degradation during interphase
APC/C
E3 ubiquitin ligase composed for multiple subunits including Cdc20 or Cdh1 as co-activator molecules
Targets multiple cell cycle regulators for ubiquitin-dependent degradation during mitosis (cyclin B, securin) or the mitotic/interphase transition (e.g., Aurora A, Plk1, or Tpx2)
SPINDLE REGULATORS
Kinesins
More than 600 proteins including Eg5, CenpE, MCAK
Microtubule-based motor proteins that hydrolyze ATP to generate energy for movement along microtubule fibers
Cyclin-Dependent Kinases and Their Regulators
Retinoblastoma Proteins and E2F Transcription Factors
Cell Cycle Phosphatases
Ubiquitin-Dependent Protein Degradation
The Mitotic Spindle and Mitotic Kinases and Kinesins
Entry into the Cell Cycle
DNA Replication
Mitosis
Cell Cycle Checkpoints
Control of the Cell Cycle