© Springer International Publishing Switzerland 2015
Varsha Gandhi, Kapil Mehta, Rajesh Grover, Sen Pathak and Bharat B. Aggarwal (eds.)Multi-Targeted Approach to Treatment of Cancer10.1007/978-3-319-12253-3_33. Chronic Lymphocytic Leukemia at the Genomic Level
(1)
Department of Experimental Therapeutics, University of Texas, M.D. Anderson Cancer Center, 301429, Houston, Texas, USA
(2)
Graduate School of Biomedical Sciences, The University of Texas, Houston, TX, USA
(3)
Department of Leukemia, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Abstract
Chronic lymphocytic leukemia (CLL), a cancer of the B lymphocytes, commonly occurs in adults in Western countries. The Rai and Binet staging systems were first employed to classify CLL based on its progression. Later, immunoglobulin variable region (IgVH) mutation status and ZAP-70 status were used as prognostic markers to indicate disease severity. Cytogenetic analyses have identified many chromosomal abnormalities that play a major role in determining the progression and outcome of CLL. Some frequently occurring abnormalities are 17p, 11q, and 13q chromosomal deletions. These factors all play a critical role in determining not only life expectancy but also the therapy course for patients. However, CLL is a very heterogeneous disease and requires deeper insight at the molecular and genetic levels to predict disease outcome. The advent of whole-genome sequencing and whole-exome sequencing ushers in a new era in the treatment of CLL. This chapter will summarize the recent major findings in the literature, including consideration of gene mutations and deletions and their effects on therapy. These genetic profiles may prove useful as potential targets for personalized therapy and better cure rates.
Keywords
Chronic Lymphocytic LeukemiaCLLGenomeGeneticsMutationsCytogeneticsNOTCH1SF3B1BIRC3XPO1WGSChronic lymphocytic leukemia (CLL), a disease of the B lymphocytes, is the most common leukemia in adults in the Western world. Cases of CLL exhibit a heterogeneous clinical course, ranging from an indolent disorder with a normal life span to an aggressive disease with short survival. Both the aggressive and indolent forms of CLL show clonal expansion of CD5-positive B cells.
Since the initial identification of CLL, scientists and clinical researchers have sought to recognize the inherent properties of the disease at the genetic level. Earlier work established disease staging such as the Rai and Binet systems that depends on the bulkiness of the disease and dissemination to other organs. This was followed by cytogenetic classification of the disease. Additional studies identified ZAP-70 and IgVH mutational status as important factors, and in the last few years, whole-genome sequencing (WGS) and whole-exome sequencing (WES) have defined new characteristics of this disease and their role in pathogenesis, prognosis, and treatment. This chapter summarizes earlier staging systems and our present knowledge of CLL at the genomic level.
3.1 Rai and Binet Staging
Two staging systems are used to classify CLL, based on disease progression. Rai staging is used in the United States and is based mainly on lymphocytosis. In the Binet system, used in the United Kingdom and Europe, CLL is classified by the number of affected lymphoid tissues. These staging systems cannot be used to predict the individual risk of disease progression and survival in the early stages of CLL. In contrast to other types of leukemias, treatment of CLL is initiated not upon diagnosis but upon progression to symptomatic disease. Patients with early Rai or Binet stage disease have a greater than 10-year life expectancy, whereas patients with advanced, fludarabine-refractory disease have a median survival of approximately 1 year (Brown 2011).
3.2 IgVH Gene Mutation and ZAP-70 Status
3.2.1 IgVH Mutation Status
The clinically distinct CLL subtypes are characterized by high and low numbers of somatic hypermutations in the variable region of the immunoglobulin genes. CLL is divided into two main subgroups based on the presence or absence of acquired somatic mutations in the immunoglobulin heavy chain gene (IGHV) expressed by leukemic B cells. Patients whose tumor cells express an IGHV gene carrying somatic mutations (M-CLL) have a more indolent disease and longer overall survival than do patients whose tumors express an IGHV gene in the germ line or unmutated configuration (UM-CLL). In the seminal paper by Hamblin et al., the median survival was 117 months for patients with UM-CLL compared with 293 months for patients with M-CLL (Hamblin et al. 1999). IgVH mutation status had prognostic significance even when consideration was restricted to patients with early stage (Binet stage A) CLL: median survival durations were 95 months (UM-CLL) and 293 months (M-CLL). Oscier et al. have confirmed the prognostic significance of IgVH mutation in multivariate analyses and additionally have shown that the presence of unmutated IgVH genes was significantly associated with deletion of chromosome 11q23, absence of deletion of chromosome 13q14, atypical lymphocyte morphology, and more than 30 % CD38 expression (Oscier et al. 2002).
Patients with UM-CLL tended to have high-risk genomic aberrations such as del17p and del11q, whereas favorable aberrations such as del13q were overrepresented in the M-CLL group. Patients with UM-CLL were also more likely to develop additional karyotypic changes (clonal evolution). Krober et al. demonstrated striking differences in the incidence of prognostically important categories between the two groups, with unfavorable cytogenetic abnormalities (17p and 11q deletions) being found almost exclusively in cases with UM-CLL (Krober et al. 2002). Interestingly, even within the UM-CLL group, those cases with high-risk genomic aberrations (as defined by 17p and/or 11q deletion) had significantly inferior survival.
3.2.2 ZAP-70 Status
Another prognostic feature, ZAP-70, was identified only a decade ago (Rassenti et al. 2004). ZAP-70 belongs to a family of protein tyrosine kinases closely related to src and is one of the membrane components associated with early cell activation in T lymphocytes and natural killer (NK) lymphocytes (Mustelin and Tasken 2003). Rassenti et al. found that patients with 20–30 % of ZAP-70 positivity were above the threshold and had a shorter life span (Rassenti et al. 2004). They reported that the median time from diagnosis to initial treatment among patients with ZAP-70-positive CLL cells that expressed an unmutated IgVH was 2.8 years, which was similar to the 4.2 years for patients with ZAP-70-positive CLL cells that expressed a mutated IgVH gene. Since their study involved a large pool of 307 patients, they observed discordance between ZAP-70 and IgVH mutational status. Further studies using microarray experiments found that ZAP-70 was differentially expressed in UM-CLL and M-CLL, and its expression has been validated as a powerful predictor of unmutated IgVH status, rapid disease progression, and inferior survival (Orchard et al. 2004).
3.3 Cytogenetic Abnormalities
CLL harbors chromosomal aberrations that are mostly deletions, as summarized below.
3.3.1 13q14 Deletion
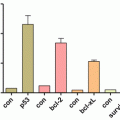
The most frequently deleted genomic region in CLL occurs at chromosome 13q14 and is associated with the indolent form of the disease. Two microRNA genes, mir-15a and mir-16-1, which are located in the crucial 13q14 region, have been implicated in CLL pathogenesis. CLLs that have a 13q14 deletion as the sole abnormality have a favorable disease course. Detailed deletion and expression analysis showed that miR15 and miR16 are located within a 30-kb region of loss in CLL and that both genes are deleted or downregulated in most CLL cases (approximately 68 %). The presence of both clonal homozygous and heterozygous deletions and the very high frequency of 13q14 loss suggest that these deletions are of pathogenic significance. BCL-2 is a target of miR15/16 and is a critical oncogene in a number of hematological malignancies as well as in solid tumors. It functions by promoting survival and inhibiting cell death. A high-resolution map of 13q14 deletions in 171 CLL samples indicated that this region also contains the DLEU7 gene, which was previously identified as a candidate tumor-suppressor gene located telomeric to miR15/16. Importantly, compared with normal B cells, malignant CLL lymphocytes showed lower expression of DELU7 (Pekarsky and Croce 2014). The 13q14 region of homozygous loss is of particular interest because it contains at least four noncoding genes including the two miR15/16 and Leu-1 and Leu-2 (Calin et al. 2002). Noncoding RNAs have roles in a great variety of processes, including transcription and chromosome structure, RNA processing and modifications, mRNA stability and translation, and protein stability and transport. Therefore, it is possible that the CLL gene(s) on 13q14 acts in a different way compared with classic tumor-suppressor genes.
3.3.2 17p Deletion
Deletion of 17p involves the loss of the TP53 tumor-suppressor gene and is found in only 5–7 % of patients with early stage CLL, but the incidence increases to 25–40 % in patients with advanced refractory disease (Fink et al. 2006), and 17p deletion constitutes a poor prognostic factor (Trojani et al. 2010). Interphase fluorescence in situ hybridization (I-FISH) was used to characterize recurrent CLL-related chromosomal abnormalities, and this analysis clearly showed that patients with deletions of 17p13 containing the TP53 locus (del(17p)) had a significantly shorter median overall survival (32 months) than did patients with normal karyotype (111 months) and also compared with patients with other recurrent chromosomal aberrations (79 months for deletion 11q, 114 months for trisomy 12, 133 months for deletion 13q) (Dohner et al. 2000). The highest incidence of TP53 mutation is seen in patients with fludarabine-refractory CLL, and much of the heterogeneity in mutation prevalence is explained by different patient cohorts. The European Research Initiative on CLL (ERIC) has compiled a large set of TP53 mutations from CLL samples and compared this profile with the currently available TP53 mutation databases (Zenz et al. 2010). From four different cohorts of CLL patients, they demonstrated that the TP53 mutation profile is independent of 17p deletion or previous therapy. These findings suggest that the TP53 mutation (detectable after therapy) is selected rather than being caused by, for example, alkylating agents. These findings also support the growing evidence that the clinical consequences of 17p deletion (and TP53 mutation) are very similar to those for the TP53 mutation in the absence of 17p. They found codon 209 frameshift mutation to be a “hot spot” for CLL.
3.3.3 11q Deletion
Deletion of 11q involves the loss of the ATM gene, which phosphorylates p53 upon DNA damage. This deletion is found in 10–20 % of CLL patients and confers an impaired clinical outcome, and mutations in the remaining allele confer a worse clinical prognosis. The I-FISH study showed that 11q deletion is the second most common aberration in CLL, occurring in 43 (20 %) of 214 cases as shown by use of the YAC probe for 11q22.3-23.1. Deletions were associated with extensive peripheral, abdominal, and mediastinal lymphadenopathy, and patients with 11q deletions had more rapid disease progression. The prognostic effect on survival strongly depended on the age of the patients. In patients less than 55 years old, median survival time was significantly shorter among patients with deletion, whereas patients older than 55 years had no significant difference compared with such patients with other chromosomal changes. Mutations can be present in the germ line, suggesting that individuals heterozygous for ATM mutations may be predisposed to develop CLL. Recently, loss of distal 11q (which includes the ATM gene) was associated with DNA repair deficiency and increased sensitivity to ionizing radiation (Parikh et al. 2007). BIRC3 mutations were recently identified in CLL and may be important for elucidating the molecular genetics of 11q22-q23 deletion (Rossi et al. 2012a).
Mutations in ATM predispose patients to lymphoid malignancies. Using FISH, a large series (more than 200 B-CLLs) found that deletion of the chromosomal region 11q22-q23 is a recurrent aberration in B-CLL (Dohner et al. 1997). Loss of the region was shown to be a prognostic marker predicting poor survival. The commonly deleted region was defined as a 2–3 mega-base-pair (Mbp) segment containing ATM, RDX, FRDX1, RAB39, CUL5, ACAT1, NPAT, KDELC2, EXPH5, MRE11, H2AX, and BIRC3. The size of the 11q deletion seems to vary between tumors but generally can be classified either as a large 11q deletion (more frequent) or a small 11q deletion (rare). Although the prognostic significance of these differences is unclear at present, the smaller deletions tend to coincide with mutations in the ATM gene, and it has been proposed that haploinsufficiency of a larger number of genes on chromosome 11q may provide a proliferative advantage comparable to biallelic inactivation of ATM (Gardiner et al. 2012). CLL subsets with 11q deletion are associated with an elevation of gene copy number alterations, representing genomic instability. Dohner and coworkers reported a median survival of 79 months in 56 patients with an 11q deletion (Dohner et al. 1997). Catovsky and coworkers found that an 11q deletion was associated with a complete response plus nodular partial response rate of 28 % and that in addition to 17p deletion and unmutated IGHV genes, 11q deletion was also an independent factor predicting shorter progression-free survival and poorer response to treatment (Catovsky 2007). In a study conducted at MD Anderson Cancer Center, CLL with an 11q22 deletion was associated with prolonged survival (97 % and 91 % at 1 and 3 years, respectively) in all patients studied and with high rates of response and relapse-free survival in patients who required therapy. In particular, regimens containing fludarabine-cyclophosphamide-rituximab (FCR) resulted in a response rate of 100 % (complete response, 76 %). In all patients, the relapse-free survival rate was 77 % at 3 years (Tsimberidou et al. 2009).
A recent study that analyzed the ATM coding region in 318 patients with CLL (140 with a chromosome 11q deletion and 178 with no 11q deletion) and 281 controls found that, compared with healthy individuals, constitutional pathogenic ATM mutations were increased in patients with chromosome 11q deletions but not in those without 11q deletions. These results suggest that ATM germ line heterozygosity does not play a role in CLL initiation but rather influences rapid disease progression through ATM loss (Skowronska et al. 2012).
Saiya-Cork and colleagues found that the insulin receptor (INSR) was significantly overexpressed in about 25 % of CLL cases, many of which carried 11q deletion (Saiya-Cork et al. 2011). INSR expression varied across a large cohort of CLLs but was overexpressed in 11q subset. Approximately two thirds of the deletion 11q CLLs showed elevated INSR expression. This receptor was functional, in that insulin was able to induce activation of the AKT and RAS/RAF/extracellular signal-regulated kinase (ERK) pathways and tyrosine phosphorylation of IRS1. These signals provide survival support to the CLLs, which show a reduction in apoptosis in response to insulin.
3.3.4 Trisomy 12
Trisomy 12 is among the most common aberrations in CLL (10–20 %), but the genes involved in the pathogenesis of CLL with trisomy 12 are unknown (Crossen and Horn 1987). Despite the frequency of trisomy in CLL, little is known about its origin, pathogenic significance, or the mechanism by which it may contribute to malignant transformation or disease progression. Through restriction fragment length polymorphism analysis, the extra chromosome 12 has been shown to derive from duplication of one chromosome 12, with retention of the other homologue rather than from triplication of one homologue (Einhorn et al. 1989). Trisomy 12 can be detected in about one third of CLL patients with clonal chromosome abnormalities and occurs as a single aberration in more than half of them. The presence of trisomy 12 is an adverse prognostic factor and predicts a short treatment-free survival.
At present, the biological significance of this different distribution of +12 cells in CLL is unknown, as is the role (if any) of this trisomy in the pathogenesis of lymphoproliferative disorders. A study of adhesion molecules in B-CLL cells, simultaneously evaluated in the peripheral blood, bone marrow, and lymph nodes, has shown that CD44 and CD54 on CD5+ CD23+ lymphocytes were more dominant in more numbers of cells in the lymph nodes than in the peripheral blood or bone marrow. In some patients with chronic lymphoproliferative diseases other than CLL, polysomy 12 detected in lymph node cells was clearly associated with progressive disease (Callea et al. 1996; Younes et al. 1994).
While not that common, trisomy 19 has also been identified in CLL (Schwaenen et al. 2004). Schwaenen and colleagues investigated 106 B-CLL cases by array comparative genomic hybridization (CGH) and found trisomy 19 as a chromosomal aberration in almost 5 % of cases (Schwaenen et al. 2004). Interestingly, this aberration seemed to be correlated with trisomy 12 and mutated IGHV genes. A similar incidence was also observed in another study of 125 B-CLL cases (Dicker et al. 2006).
3.4 Genomics of CLL
Recent advances in DNA sequencing technology have facilitated the analysis of entire genomes of individual cancers and have led to the identification of novel genetic alterations. Although the causative somatic genetic mutations for CLL have not yet been defined, massively parallel sequencing technology has led to the discovery of several genes that are mutated in CLL. Although some of these genes were identified previously, such as p53 and ATM, new genes (albeit mutated in a smaller fraction of the patient population) have been identified by recent sequencing endeavors.
3.4.1 WGS and WES
The Spanish CLL Genome Consortium performed the first sequencing in CLL. This study by Puente et al. reported the WGS of two cases of CLL without mutations in immunoglobulin genes (IGHV unmutated) and two with mutations in immunoglobulin genes (IGHV mutated) (Puente et al. 2011). Whole-genome sequencing of the tumors using germ line DNA as a baseline identified approximately 1,000 somatic mutations in each of the tumors, consistent with prior estimates of one mutation per Mb for leukemias. From all of the somatic mutations identified in the above four patients, 46 mutations were predicted to alter the protein coding sequence of 45 genes, of which 26 were found to be expressed at the RNA level and of potential biological relevance. Sanger sequencing in a validation cohort of 169 additional CLL patients confirmed that four of these genes (NOTCH1, MYD88, XPO1, and KLHL6) were recurrently mutated.
3.4.1.1 NOTCH1
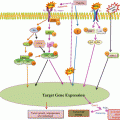
NOTCH1 is a member of an evolutionally conserved signaling pathway with a pivotal role in numerous important developmental and physiological processes. NOTCH1 has been sporadically reported as one of the genes functionally mutated in CLL (Sportoletti et al. 2010). In genome-sequencing efforts, NOTCH1 was among the top genes identified as having activating alterations among CLL patients (Puente et al. 2011; Fabbri et al. 2011; Wang et al. 2011). Overall, fewer than 10 % of patients are reported with this anomaly, but among patients with a poor prognosis, such as those with unmutated IgVH and ZAP-70+, the incidence increases (Puente et al. 2011). NOTCH1 mutations have been identified particularly in patients with trisomy 12 (Fabbri et al. 2011; Balatti et al. 2012; Del Giudice et al. 2012). Importantly, in chemorefractory disease and during disease progression toward Richter’s transformation, the incidences increase to 21 % and 31 %, respectively (Fabbri et al. 2011). Activation of NOTCH signaling is not exclusive to CLL; in fact, this gene is among the highly mutated genes in acute lymphoblastic leukemia (O’Neil et al. 2007; Weng et al. 2004). Because FBXW7 targets activated NOTCH1 for degradation, FBXW7 mutation might be a second mechanism leading to activated NOTCH signaling (Jeromin et al. 2014).
NOTCH1 mutations have proven to be independent markers of shorter survival (Rossi et al. 2012b). Moreover, these mutations have been associated with a significantly shorter treatment-free survival and overall survival (Rossi et al. 2012a; Puente et al. 2011). Mechanistically, both the canonical and noncanonical pathways of NOTCH signaling are constitutively upregulated in NOTCH1-mutated patients, irrespective of the levels of expression of the receptor and of presenilin1/2, the catalytic subunits of the enzyme that controls NOTCH1 final activation (Arruga et al. 2014). Gamma secretase inhibitors as well as antibodies against NOTCH1 are currently available for testing in a preclinical setting (Aster and Blacklow 2012; Gounder and Schwartz 2012; Krop et al. 2012; Tolcher et al. 2012).
3.4.1.2 SF3B1
An additional pathway that has several genes mutated is RNA splicing and processing. The primary locus in this pathway is the splicing factor SF3B subunit 1 (SF3B1) gene. SF3B is a U2 small nuclear ribonucleic particles (snRNP) to the branch point of 39 splicing sites associated protein complex essential for spliceosome assembly (Corrionero et al. 2011; Wahl et al. 2009). This factor is phosphorylated by DYRK1A protein kinase (de Graaf et al. 2006).
SF3B1 was found independently by several investigators to be mutated in CLL (Wang et al. 2011; Rossi et al. 2012b; Quesada et al. 2012) and other hematological malignancies (Papaemmanuil et al. 2011; Visconte et al. 2012). SF3B1 mutations were detected at frequencies ranging from 5 % to 10 % in patients at diagnosis, with a sharp increase in frequency (17–24 %) among patients with progressive, refractory disease and poor outcome. In CLL, SF3B1 mutations are associated with faster disease progression and poor overall survival (Quesada et al. 2012). This gene alteration was associated with shorter time to treatment (Wang et al. 2011; Quesada et al. 2012; Mori et al. 2012) as well as fludarabine refractoriness (Rossi et al. 2011). The enhanced expression of the putative splice junctions that lead to the truncated form of ATM in the SF3B1-mutated cells was identified using gene annotation enrichment analysis (Arruga et al. 2014). SF3B1 mutations tend to be mutually exclusive with ATM mutations and 11q deletions, supporting the recently proposed connection between mutations in SF3B1 and ATM dysfunction (Wang et al. 2011; te Raa D et al (2012). SF3B1 mutations were associated with CD38 positivity, advanced disease stage, and unmutated IGHV. Although functional consequences of SF3B1 mutations have not yet been defined, these anomalies are supposed to lead to a modified function of SF3B1, possibly due to altered interaction with other proteins (Jeromin et al. 2014). An inhibitor of this factor, spliceostatin A, is responsible for nuclear retention of pre-mRNA (Tolcher et al. 2012).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree