Fig. 2.1
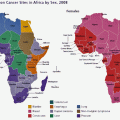
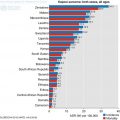
The age standardised incidence rate of the most common cancers in (a) females and (b) males in SSA (Source: GLOBOCAN 2012 (IARC))
2.1.1 Environmental Factors
Inter and intra-regional variations are mostly due to environment factors. The environment plays an important role in carcinogenesis and it is postulated that it contributes about 70% to lifetime risks of most cancers (Wu et al. 2016). In SSA, 32.7% of cancers are attributable to infection compared to 7.4% in more developed countries (De Martel et al. 2012; Okuku et al. 2013). Hepatitis B and C viruses (HBV, HCV), Human papilloma virus (HPV), Helicobacter pylori, Epstein-Barr virus (EPV) and Human herpes virus (HHV) type B accounted for over 97% of all infected related cancer in 2008 (De Martel et al. 2012). There are however other environmental factors involved in carcinogenesis with differing relative contributions (Table 2.1). These environmental factors may lead to extrinsic mutational signatures (Wu et al. 2016).
Cancer types | Extrinsic risk (%) | Examples of potential extrinsic risk factors |
|---|---|---|
Breast | Substantial | Oral contraceptive, hormone replacement therapy, lifestyle (diet, smoking, alcohol, weight) |
Prostate | Substantial | Diet, obesity, smoking |
Lung | >90 | Smoking, air pollutant |
Colorectal | >75 | Diet, smoking, alcohol, obesity |
Melanoma | 65–86 | Sun exposure |
Basal cell | ~90 | UV |
Hepatocellular | ~80 | HBV, HCV |
Gastric | 65–80 | H. pylori |
Cervical | ~90 | HPV |
Head & neck | ~75 | Tobacco, alcohol |
Esophageal | >75 | Smoking, alcohol, obesity, diet |
Oropharyngeal | ~70 | HPV |
Thyroid | >72 | Diet low in iodine, radiation |
Kidney | >58 | Smoking, obesity, workplace exposures |
Thymus | >77 | Largely unclear |
Small intestine | >61 | Diet, smoking, alcohol |
Extranodal non-Hodgkin’s lymphoma (NHL) | >71 | Chemicals, radiation, immune system deficiency |
Testis | >45 | Largely unclear |
Anal and anorectal cancers | >63 | HPV, smoking |
2.1.2 Mutational Signatures
With many cancers, there is an accumulation of an average of 90 mutant genes but only a subset contribute to neoplastic process, with each cancer having its own distinct mutational signature (Sjöblom et al. 2006). Analysis of these mutational signatures, the fingerprints left on cancer genomes by different mutagenic processes, showed that intrinsic cancer mutations (random errors in DNA replication) showed strong positive correlations with age suggesting acquisition at a relative constant rate over lifetime. However, it is postulated that all other mutational signatures that lack consistent correlations with age, suggests acquisition at different rates in life and are thus likely a consequence of extrinsic carcinogen exposure (Wu et al. 2016). Cancers that have substantial environmental risk proportions (Table 2.1) harbour large percentages of extrinsic mutational signatures (environmental factors that affect mutagenesis rate), for example approximately, 100% for myeloma, lung, and thyroid cancers, and approximately 80–90% for bladder, colorectal and uterine cancers (Wu et al. 2016).
These extrinsic mutational signatures are most likely to partly explain the geographical variation in cancer epidemiology in terms of incidence and biology. Amongst migrants moving from low-risk to high-risk countries, the incidence of colorectal cancer tend to increase towards that of the local population (Haggar and Boushey 2009). Apart from migration, other geographical factors come into play and that includes urban residency. Current urban residency is a stronger predictor of risk than is an urban location of birth (Haggar and Boushey 2009).
2.1.3 Global Molecular Pathways
Global molecular pathways for most cancers are well established. Geographical and/or racial differences tend to be in the prevalence of mutations in candidate genes and the effect and types of polymorphisms and epigenetic phenomena have on these genes. All these may enhance understanding of differences in tumour biology, identify tumours with poor prognosis, serve as biomarkers, and importantly, help direct appropriate therapy.
2.1.4 Genomic Research Output from Sub-Saharan Africa
Cancer genomic research output from SSA is low apart from South Africa who has taken strides in developing biotechnology industry. However, there were only 31 published research papers in cancer genomics in SSA between 2004 and 2013 (Adedokun et al. 2016). Encouragingly, with increasing collaboration between institutions in SSA countries and those in United States and Europe, cancer genomic research output is increasing. The rest of the chapter will give a selective, brief overview. It is not meant to be exhaustive but to highlight the theme as it relates to SSA.
2.2 Global Genomic Differences
Global genomic pathways for most cancers are well established. The importance of assessing differences in different geographic regions and races is to help in prognostication and/or planning treatment. Another important aspect is finding out different mutations that may affect the function of the affected gene. Two common cancers are used to illustrate this.
2.2.1 Breast Cancer
BRCA1 and BRCA2 are the two most commonly mutated tumour suppressor genes associated with early onset and familial breast cancer (Fackenthal et al. 2012). The prevalence of BRCA1 and BRCA2 mutations is higher in Nigerians (7.1% and 3.9% respectively) than African-Americans (1.4% and 2.6% respectively). BRCA1 prevalence in Nigerians (7.1%) is also higher than the 2.9% in Caucasian Americans (Fackenthal et al. 2012). Importance of this is that more than 50% of women with BRCA1 mutation have a chance of developing triple negative breast cancer (Brewster et al. 2014). BRCA1 tumours have no defined therapeutic target and can be aggressive when refractory to current treatment. BRCA2 mutation carriers, on the other hand, are usually oestrogen receptor (ER) positive, and they are amenable to hormonal treatment (Fackenthal et al. 2012).
Triple negative breast cancer (TNBC) are oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) negative. This means that TNBC negative patients do not respond to ER modulators (e.g. tamoxifen) or aromatase inhibitors (anastrazole), and for those who are HER2 negative, they do not respond to trastuzumab, a monoclonal antibody. TNBC tumours are aggressive, and chemotherapy is the standard treatment in adjuvant or neo-adjuvant setting with a complete response rate of 30–45% (Brewster et al. 2014). Of note, ER-positive breast cancer in blacks living in the United States and United Kingdom is between 61% and 66% compared to 25% in Nigerians and Senegalese (Huo et al. 2009).
Fackenthal et al. (2012) sequenced the whole of BRCA1 and BRCA2 genes and found that Nigerian breast cancers had 11% mutation rate in BRCA1/2 genes which they wrote, was larger than any reported for a non-founder unselected population. 62.5% of the distinct BRCA1/2 mutant alleles identified were recurring mutations, occurring in two or more subjects. The authors suggested that BRCA1/2 mutation frequencies in Nigerians could be an underestimate (Fackenthal et al. 2012).
2.2.2 Colorectal Cancer
In colorectal cancer (CRC) patients, there are less frequent mutations of K-RAS (21–32%) and BRAF (0–4%) in Ghanaian and Nigerian patients (Abdulkareem et al. 2012; Raskin et al. 2013) compared to approximately 40% K-ras and 10–15% BRAF mutations in Europeans (Sforza et al. 2016). This is important because mutation of K-RAS and BRAF, which are downstream of epidermal growth factor receptor (EGFR), cause persistent activation of downstream signalling regardless of EGFR inhibition by monoclonal antibodies, cetuximab and panitumumab (Sforza et al. 2016).
However, in the Nigerian CRC analysis by Abdulkareem et al. (2012), only K-RAS codons 12, 13 and 61 and BRAF codon 600 were assessed and in the Ghanaian CRC cohort, only KRAS exons 2 (codon 12, 13) and 3 (codon 61) and BRAF exon 15 were analysed (Raskin et al. 2013). It is therefore possible that there could be other mutations in the non-sequenced exons that could be deleterious. Low or no BRAF mutations in the presence of MSI-H raises the prospect of Lynch syndrome (Setaffy and Langner 2015) but this is a rarity in SSA (Irabor and Adedeji 2009) or possibly unrecognised. Patients with KRAS and BRAF mutations also have poorer disease free and overall survival compared patients without mutations in these genes (Modest et al. 2016). Also, allele specific inhibitors may be therapeutic options in future (Lito et al. 2016).
Microsatellite instability high (MSI-H) tumours in Ghanaians were 41% compared to 15–45% of African-Americans and 10% whites, but MSI-low tumours were 20% in Ghanaians compare to 4–5% of African-Americans while microsatellite stable (MSS) tumours were found in 39% of Ghanaians in 50–85% of African-Americans with colorectal cancer (Raskin et al. 2013). MSI-H colorectal patients have a less aggressive clinical behaviour and a favourable prognosis compared to MSS patients. There is however conflicting evidence about the predictive role of MSI regarding response to 5-FU based adjuvant chemotherapy (Saridaki et al. 2014).
2.2.3 Comments
More work is needed in the established molecular pathways of common cancers in SSA. Using standard European or American genetic panels may not recognise variants and polymorphisms in common cancer genes in SSA. This may underestimate non-synonymous mutations. It may be that while trying to determine molecular basis of phenotypic differences, whole genes of interest should be sequenced rather than selective codons based on panels currently available in the West.
2.3 Single Nucleotide Polymorphism (SNP)
Another important aspect in cancer pathway are genetic variants, and these include single nucleotide polymorphism (SNP) and copy number variation (CNV). They help in determining susceptibility to certain type of cancers (Chung et al. 2010; Redon et al. 2006). SNPs are single nucleotide base substitution that affects at least 1% of the population. The important ones alter coding sequence and result in a non-synonymous change, a shift in the amino acid sequence of protein that may change its function (Chung et al. 2010).
Genome wide association studies (GWAS) with SNPs investigate mainly susceptibility to cancer and outcome. Outcome studies help to determine prognostic information for survival, complications or response to pharmacological interventions (Erichsen and Chanock 2004). While there is plethora of GWAS, there is growing evidence that these may be different within racial groups and between geographical regions (Cook et al. 2014; Murphy et al. 2012).
2.3.1 Prostate Cancer
GWAS of prostate cancer in Ghanaians showed evidence of a new SNP locus at chromosome (Chr) 10p14 associated with prostate cancer and another SNP at Chr. 5q31.3 associated with high Gleason score of 7 (Cook et al. 2014). Its role in pathogenesis is yet unclear. Most importantly however was that, of the 81 previously reported prostate cancer susceptibility loci, only 10 SNPs (including 4 SNPs in regions 1 and 2 of Chr. 8q24) were statistically significant in Ghanaians. These previously reported SNPs were mostly from European, Asian and African-American ancestry. The authors postulated that the differences may be due to distinct genomic architecture, heterogeneous prostate cancer populations or yet uncharacterised gene-environment interactions (Cook et al. 2014).
Another study tested for ten SNPs from 4 regions of Chr. 8q24 in 1157 Nigerian (West Africa), Cameroonian (West Africa) and Jamaican (Caribbean) patients with prostatic cancer (Murphy et al. 2012). Four SNPs were significantly associated with prostatic cancer in the West Africans but there were no SNP associations observed in the Jamaican population. Both studies above show a geographical variation to genomic variants associated with disease, and they may lead to better understanding of pathogenesis. More importantly, they may act as biomarkers for early detection specific to a region or race (Murphy et al. 2012).
Environment alone does not answer the differences in SNPs between races. Assessment of 12 SNPs associated with oesophageal squamous cell carcinoma in eight genes between black and mixed ancestry South Africans. There were no observed associations in black South Africans, while there were several significant or suggestive associations in mixed ancestry South Africans (Bye et al. 2011). Interestingly, many of the associations in the mixed ancestry South Africans were previously reported in Europeans and Asians.
2.3.2 Race and Migration
Apart from geographical variation in SNP association with disease, this association may change with migration. Assessment of allele frequency of hepatocellular cell carcinoma (HCC) associated SNPs in 53 human populations showed differences in SNP expression and susceptibility to HCC (Ngamruengphong and Patel 2014). The genetic risk score for HCC was calculated based on combined risk of two SNPs associated with a moderate risk of HCC in patients with chronic HCV and HBV. The risk associated with the SNPs was high in populations from Africa and decreased with migration to Europe and Central Asia (Ngamruengphong and Patel 2014).
There are accumulating GWAS involving Africans but what they lack is consistency and validation. Translational research is essential to assess clinical significance and more importantly in poorly resourced countries, cost-benefit studies. What is also becoming clear is that many GWAS are probably more regional based more so than global genomic pathways.
2.3.3 Variants in Infective Organisms
As a third of cancers in SSA are infection related, variants in infective organisms are also important. Papillomaviruses are small double stranded DNA viruses, species specific. They are of various types defined by nucleotide sequences of the L1 open reading frame. Each type acquire SNPs and/or insertion/deletions (indels) which tend to become fixed within viral linkages and overtime, the quantity of variants increase leading to speciation or viral lineages (Burk et al. 2013).
A study 530 HPV6 DNA-positive samples from 15 countries across six continents collected from anogenital, head and neck regions were analysed (Jelen et al. 2014). There were two distinct viral lineages A and B, five B sub-lineages. Table 2.2 shows the distribution of lineage A and sub-lineages B according to geographical location, anatomical location, type of lesion and gender. Lineage B was most prevalent lineage worldwide apart from Asia, where lineage A predominates. Sub-lineage B3 was the most prevalent in South Africa. B2 was found in all continents except Asia, B4 was found only in Europe and Asia and B5 was found only in Europe and Africa (Jelen et al. 2014). Sub-lineage B3 was found in less than 5% of ano-genital wart tissue. The importance of these differences is the provision of valuable resource for functional pathogenicity, vaccination and molecular assay development (Jelen et al. 2014) that can be specific for different environments.
Table 2.2
HPV6 lineage A and sub-lineage B according to geographical location, anatomical location of infection, lesion type and gender (Jelen et al. 2014).
Variable | No (%) of samples | ||||||
|---|---|---|---|---|---|---|---|
A | B1 | B2 | B3 | B4 | B5 | Total | |
Geographical location | |||||||
Europe | |||||||
Croatia | 6 (12.8) | 34 (72.3) | 2 (4.3) | 5 (10.6) | 0 (0) | 0 (0) | 47 (100) |
Czech Republic | 17 (32.1) | 32 (60.4) | 2 (3.8) | 1 (1.9) | 1 (1.9) | 0 (0) | 53 (100) |
Germany | 2 (4.2) | 26 (54.2) | 11 (22.9) | 9 (18.8) | 0 (0) | 0 (0) | 48 (100) |
Lithuania | 6 (85.7) | 1 (14.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (100) |
Serbia
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

| |||||||