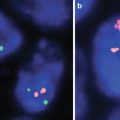
Fig. 9.1
ESR1 (ER) positive by immunohistochemistry
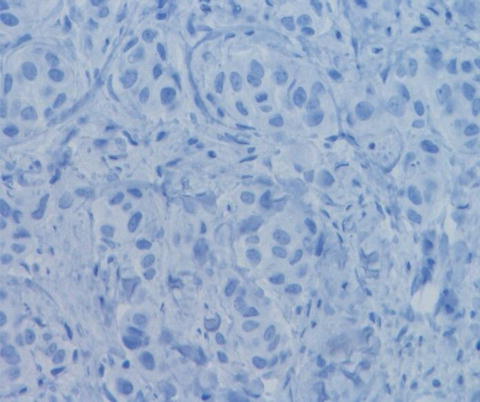
Fig. 9.2
ESR1 (ER) negative by immunohistochemistry
The IHC results can be reported either as percentage of positive cells or by using one of the two HR quantifying systems: Allred score (Table 9.1) [23] and H score (Table 9.2) [24].
Table 9.1
Allred score—Total score = PS + IS (adapted after Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999 May; 17(5):1474–81)
(%) Immunoreactivity of tumor cells | Proportion score (PS) |
0 | 0 |
<1 % | 1 |
1–10 % | 2 |
11–33 % | 3 |
34–66 % | 4 |
≥67 % | 5 |
Staining intensity of tumor cells | Intensity score (IS) |
Absent | 0 |
Weak | 1 |
Intermediate | 2 |
Strong | 3 |
Table 9.2
H score (adapted after McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS S. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985 Aug; 109(8):716–21)
Staining intensity | Value × corresponding % of tumor cells |
|---|---|
Absent | 0 |
Weak | 1 × % |
Intermediate | 2 × % |
Strong | 3 × % |
Score | =sum of individual values |
The Allred score combines the percentage of positive (nuclear staining) tumor cells (proportion score) with the staining intensity (intensity score) into a total score (=PS + IS). An Allred score of 0–2 should be reported as negative and of 3–8 as positive (Table 9.1) [15, 23].
Similarly, the H score combines the percentage of positive tumor cells with the staining intensity. The final score represents a sum of each staining intensity (scored 0–3) multiplied with the corresponding percentage of positive tumor cells. An H score of <1 should be reported as negative (Table 9.2) [24].
Approximately 70–80 % of primary breast carcinomas are ESR1 (ER) positive [15]. Of these about 70 % show response to hormonal therapy. Resistance to hormonal therapy and decreased ESR1 (ER) expression correlate with an aggressive clinical course and disease progression. Several treatment options are available for hormone-sensitive breast cancers, including the following:
Ovarian ablation can be achieved surgically by removing the ovaries but more often with the use of luteinizing hormone-releasing hormone (LHRH) analogs, such as the FDA-approved drugs goserelin (Zoladex) or leuprolide (Lupron).
Blocking estrogen production: Letrozole (Femara), anastrozole (Arimidex), and exemestane (Aromasin) are aromatase inhibitors that block estrogen production. They have been approved for the treatment of both early and advanced breast cancers [6, 8] and are only effective in women whose ovaries are suppressed [25].
Blocking estrogen’s effects: Selective estrogen receptor modulators (SERMs) bind to estrogen receptors, preventing estrogen from binding. FDA-approved SERMs are tamoxifen (Nolvadex), raloxifene (Evista), and toremifene (Fareston). Other antiestrogen drugs, such as fulvestrant (Faslodex), function as pure estrogen antagonists [6, 13, 22, 26].
The Arimidex, Tamoxifen Alone or in Combination (ATAC), double-blinded phase III clinical trial designed to evaluate and compare the ability of anastrozole and tamoxifen, alone and in combination, to prevent breast cancer recurrence in postmenopausal women with hormone receptor-positive tumors, showed that at 10 years anastrozole alone was better at preventing recurrent disease in postmenopausal women with early-stage hormone receptor-positive tumors when compared to tamoxifen alone. However, no improvement in overall survival was seen [27].
Premenopausal women with hormone receptor (HR)-positive breast cancer should be treated with 5 years of tamoxifen and postmenopausal patients with a minimum of 5 years of adjuvant therapy with an aromatase inhibitor or tamoxifen. The most recently updated guidelines recommend for pre- or perimenopausal women who have received 5 years of adjuvant tamoxifen, an extension of the treatment with tamoxifen to 10 years. Postmenopausal women who have received 5 years of adjuvant tamoxifen should either continue with tamoxifen or switch to an aromatase inhibitor for a total of 10 years of adjuvant endocrine therapy [28].
ESR1 and PGR Testing by Reverse Transcription-Polymerase Chain Reaction
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) is an alternative method for ESR1 and PGR testing. Recently, the Oncotype DX assay (discussed later) started to include in its report the ESR1 and PGR status based on quantitative measurement by RT-PCR and several studies reported good correlation with IHC testing results. Taken in consideration additional advantages of the IHC method such as testing on intact tissue fragments with preserved tumor morphology, lower cost, faster turnaround time, and availability, there is currently no definitive reason to switch to RT-PCR for ESR1 and PGR testing [29, 30].
ERBB2 (HER2) Receptor Testing
The erb-b2 receptor tyrosine kinase 2 (ERBB2, which is commonly known as HER2) is a proto-oncogene that belongs to the human epidermal growth factor receptor family of tyrosine kinases together with HER1 (EGFR), HER3, and HER4 [6, 13, 31]. The ERBB2 (HER2) gene is located on chromosome 17.9 and encodes a tyrosine-kinase receptor that is present on the surface of breast epithelium [6, 13, 32].
The epidermal growth factor receptor family are activated by ligand-induced dimerization and can homo- or heterodimerize allowing multiple receptor combinations. The formation of the dimers leads to activation of the intrinsic tyrosine kinase domain with recruitment of a variety of molecules and activation of different downstream signaling pathways, including MAPK proliferation and/or PI3K/Akt pro-survival pathways [25, 33–35].
Amplification of the ERBB2 (HER2) gene induces activation with ERBB2 (HER2) protein overexpression in 15–20 % of all breast cancers. Gene amplification correlates with protein overexpression in 95 % of cases. In the remainder of cases ERBB2 overexpression occurs through a different mechanism [6, 13].
Testing for ERBB2 (HER2) receptor status of invasive breast cancer is primarily performed to identify patients that could benefit from anti-ERBB2 therapy. Several technologies for testing (IHC, fluorescence in situ hybridization (FISH), real-time RT-PCR, and enzyme-linked immunosorbent assay (ELISA)) are available [36]. Several studies have shown a very strong correlation (in approximately 95 % of cases) between IHC ERBB2 (HER2) protein expression and gene amplification by FISH [13, 17, 37]. The current ASCO/CAP guidelines continue to endorse IHC and fluorescence ISH (FISH) techniques for ERBB2 (HER2) testing with the recent addition of “bright-field ISH,” as an alternative to FISH testing [7, 17]. Pathologists must ensure that at least one tumor sample from each invasive breast cancer (primary or metastatic) is tested either by IHC or ISH using a validated testing method.
Testing should be performed on tissue fixed in neutral-buffered 10 % formalin or another validated fixative for 6–72 h. The cold ischemia time should be as short as possible, ideally of no more than 1 h [15, 38–40]. Biopsy tissue samples fixed for less than 1 h or resection specimens fixed for less than 6 h should not be tested [16, 18].
False-positive results for ERBB2 (HER2) may be due to edge artifact, misinterpretation of cytoplasmic staining, overstaining (strong membrane staining of normal cells), or misinterpretation of carcinoma in situ as invasive tumor. Laboratory should follow the standards related to the initial test validation, ongoing internal quality assurance, external proficiency testing, and routine periodic performance monitoring. Quality assurance should include a comparison of ERBB2 IHC-positive results and amplification by FISH. The ASCO/CAP guidelines require a concordance of 95 % between IHC samples and a validated assay. External proficiency testing surveys for ERBB2 are available from CAP and other organizations to ensure that the laboratory assays are working as expected [16, 18].
ERBB2 (HER2) Testing by Immunohistochemistry
ERBB2 testing by IHC is a semiquantitative method that evaluates for protein expression on the surface membrane of tumor cells [8]. There are several antibodies of different IHC-based ERBB2 tests available including Pathway ERBB2 (HER2) (clone 4B5; Ventana Medical Systems Inc., Tucson, AZ, USA), HercepTest (polyclonal, Dako Denmark A/S, Glostrup, Denmark), and Oracle ERBB2 (HER2) (clone CB11; Leica Microsystems GmbH, Wetzlar, Germany) [16, 41–43]. For interpretation of ERBB2 IHC results the US Food and Drug Administration (FDA) approved a scoring system based on the level of immunostaining of the tumor cells (Table 9.3). Examples of ERBB2 IHC are illustrated in Figs. 9.3 and 9.4 [16].
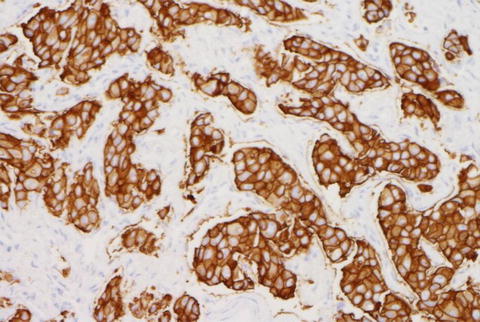
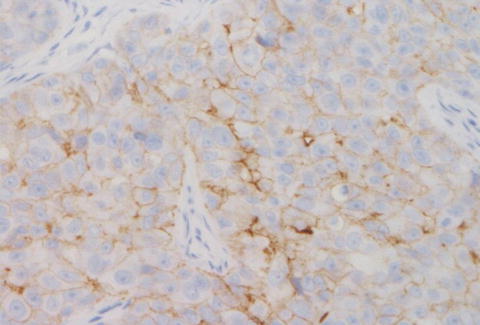
Table 9.3
ERBB2 (HER2) reporting guidelines and recommendations (adapted after Cornejo KM, Kandil D, Khan A, Cosar EF. Theranostic and molecular classification of breast cancer. Arch Pathol Lab Med. 2014 Jan; 138(1):44–56)
ERBB2 (HER2) IHC score | 0 | 1+ | 2+ | 3+ |
Result | Negative | Negative | Equivocal | Positive |
Interpretation | No staining or ≤10 % of tumor cells with incomplete faint/barely discernible membrane staining | >10 % of tumor cells with incomplete faint/barely discernible membrane staining | >10 % of the tumor cells with incomplete and/or weak to moderate circumferential membrane staining or ≤10 % of tumor cells with complete intense circumferential membrane staining | >10 % of the tumor cells with complete, intense circumferential membrane staining |

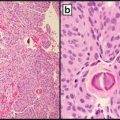
Fig. 9.3
ERBB2 (HER2) positive by IHC

Fig. 9.4
ERBB2 (HER2) negative by IHC
Results that scored equivocal as 2+ (11–30 %) should be reflexed to FISH testing (discussed below) for further confirmation [16, 18]. With a negative ERBB2 IHC result on a small biopsy sample, repeat testing on a subsequent specimen or by a different testing method should be taken into consideration, especially if the tumor shows discordant histologic features (high histologic grade, PGR negative or weakly expressed, high proliferation index).
ERBB2 (HER2) In Situ Hybridization
FISH, chromogenic in situ hybridization (CISH), and silver-enhanced in situ hybridization (SISH) are assays that can be used to determine the presence or absence of ERBB2 (HER2) gene amplification [18]. FISH detects specific DNA sequences on a chromosome using fluorescent probes. CISH technique uses a chromogen (diaminobenzidine) to create color signals that can be then identified by light microscopy on tissue section with well-preserved morphologic details. Silver in situ hybridization (SISH) uses an enzyme-linked probe causing silver ions to deposit on the target, producing a dense, high-resolution, black easily identifiable stain [16]. For additional information about ISH techniques and comparison with IHC please see Table 9.4.
Table 9.4
Comparison between ERBB2 (HER2) testing assays (adapted after Bhargava R, Esposito N, Dabbs D. Immunohistology of the breast. In: Dabbs D, editor. Diagnostic Immunohistochemistry; Theranostic and genomic applications. 3rd ed.; 2010. p. 763–819)
Pros and Cons | IHC | FISH | CISH/SISH |
Microscope | Bright field | Fluorescent | Bright field |
Tumor amount needed | Large | Small | Large |
Preservation of tumor morphology | Yes | No | Yes |
Turnaround time | 4–6 h | 3 days | 2 days (CISH) |
4–6 h (SISH) | |||
Cost and availability | Low; widely available | High; limited | Intermediate; limited |
Equivocal results | 25 % | <5 % | <5 % |
FISH for ERBB2 (HER2), the most frequently used technique, detects specific DNA sequences on chromosome 17. Some assays use a single-color probe to measure the ERBB2 gene copy number, while other assays use an additional (dual) probe (chromosome enumeration probe; CEP17) for the centromere of chromosome 17 and determine the ratio of ERBB2 signals to chromosome 17 copies. In the majority of breast carcinoma cases, both methods give the same result with rare discordances that are usually due to variation in the number of CEP17 signals. True polysomy of chromosome 17 is seen in a limited number of breast cancer cases (up to 1–2 %) [18].
Recently updated recommendations for reporting of ERBB2 testing results by ISH are presented in Table 9.5. Examples of ERBB2 FISH testing are illustrated in Figs. 9.5 and 9.6 [17, 18].
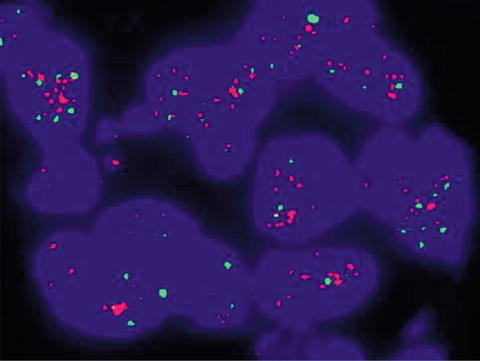
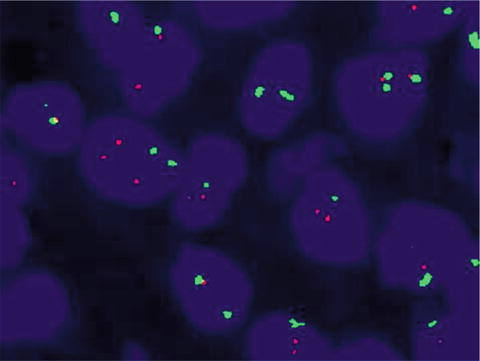
Table 9.5
ERBB2 (HER2) ISH testing result interpretation guidelines and recommendations (adapted after Cornejo KM, Kandil D, Khan A, Cosar EF. Theranostic and molecular classification of breast cancer. Arch Pathol Lab Med. 2014 Jan;138(1):44–56 and Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: ASCO/CAP clinical practice guideline update. Arch Pathol Lab Med. 2014 Feb; 138(2):241–56.)
Positive for ERBB2 (HER2) | Equivocal for ERBB2 (HER2) | Negative for ERBB2 (HER2) | Indeterminate for ERBB2 (HER2) |
|---|---|---|---|
Single-probe ERBB2 copy number ≥6.0 signals/cell | Single-probe ERBB2 copy number ≥4.0 and <6.0 signals/cell | Single-probe ERBB2 copy number <4.0 signals/cell | Inadequate handling of the tissue |
Artifacts impeding interpretation | |||
Dual-probe ERBB2/CEP17 ≥2 with average ERBB2 copy number ≥4.0 signals/cell | Dual-probe ERBB2/CEP17 <2 with average ERBB2 copy number ≥4.0 and <6.0 signals/cell ↓ | Dual-probe ERBB2/CEP17 <2 with average ERBB2 copy number <4.0 signals/cell | Analytic testing failure ↓ |
Dual-probe ERBB2/CEP17 ≥2 with average ERBB2 copy number <4.0 signals/cell | Reflex testing approximately on same specimen, IHC, or repeat ISH with chromosome 17 probe or approximately on new specimen order new test (IHC/ISH) | Retest with an alternative method or on another tumor tissue block or specimen, if available | |
Dual-probe ERBB2/CEP17 <2 with average ERBB2 copy number ≥6.0 signals/cell |

Fig. 9.5
ERBB2 (HER2) positive by FISH

Fig. 9.6
ERBB2 (HER2) negative by FISH
If there is discordance between the ERBB2 (HER2) results and the histopathologic features a repeat testing for ERBB2 should be ordered. ERBB2 ISH-negative results should trigger repeat testing if the breast tumor has a high histologic grade, if only a small biopsy with limited amount of tumor was initially available or if the subsequent resection specimen showed a morphologically distinct high-grade carcinoma when compared to the initial biopsy. Retesting should also be ordered for ERBB2-positive cases with discordant histology (histologic grade 1, ESR1 or PGR-positive tumors and tubular, mucinous, cribriform, or adenoid cystic variants of breast carcinomas) [44].
Tumors with ERBB2 (HER2) copy number of <4.0 signals/cell by single-probe ISH analysis should be further tested by dual ISH and reported as negative only if the dual-probe ISH analysis shows an ERBB2/CEP17 ratio of <2.0. Similarly a tumor with a ERBB2/CEP17 ratio of <2.0 should not be considered as negative unless the ERBB2 copy number is <4.0 signals/cell [18]. At the same time, tumors with low-level ERBB2 amplification (ERBB2/CEP17 ratio of ≥2.0 and an average ERBB2 copy number of <4.0 signals/cell) are considered positive for management purposes, although there is not enough evidence of a clinical benefit from targeted treatment for these cases [37, 45, 46].
An issue that can affect the interpretation of this test is the genetic heterogeneity described in 5–30 % of breast cancer tumors. Genetic heterogeneity was defined by the 2007 ASCO/CAP guidelines as having 5–50 % of invasive tumor cells with a ERBB2/CEP17 ratio greater than 2.2 or mean ERBB2 copy number >6 [16]. Chromosome 17 abnormalities can lead to alterations of the ERBB2/CEP17 ratio, potentially leading to equivocal or incorrect ISH results [41]. In these cases, the gene copy number may reflect more accurately the ERBB2 status. The recommendation for addressing ERBB2 interpretation issues resulting from genetic heterogeneity and chromosome 17 aneusomy is that a second aggregate population of >10 % of cells should be counted and reported separately [7]. In the case of a second contiguous population of ≥10 % cells with increased ERBB2 signals/cell, consisting of more than 10 % tumor cells, an additional at least 20 non-overlapping cells within this cell population should be counted separately and reported. An overall random count is not appropriate. In cases with tumor heterogeneity, biopsy samples may not be representative of the whole tumor and repeat testing on the excision specimen should be considered in order to correctly determine the percentage of amplified tumor, if present [7].
There are no clear indications of how to proceed when a population of <10 % is present.
Approximately 15–20 % of breast cancers show ERBB2 gene amplification or overexpression and are candidates for therapy targeting ERBB2 or its downstream pathways.
Another 15 % of breast tumors have low to intermediate levels of ERBB2 protein expression with roughly one-third of these cases also showing ERBB2 gene amplification. The benefit of targeted therapy in this last group of patients is still uncertain and needs to be further investigated [5].
Monoclonal antibodies and tyrosine kinase inhibitors are two main types of targeted therapies used in the treatment of ERBB2-positive breast cancer. Additionally, PARP inhibitors are being studied for the treatment of triple-negative breast cancer. Monoclonal antibodies may be used in combination with chemotherapy as adjuvant therapy.
Trastuzumab (Herceptin) is an FDA-approved monoclonal antibody targeting the ERBB2 (HER2) receptor and blocking the effects of ERBB2 (HER2) protein [47–50]. Five main studies evaluating this drug (HERA, FinHer, NSABP B-31, BCIRG006, N9831) have shown improvement in disease-free and overall survival, independent of the chemotherapy regimen or sequence of trastuzumab delivery [42].
Pertuzumab is another monoclonal antibody that may be used in combination with trastuzumab and chemotherapy to treat patients with ERBB2-positive breast cancers, specifically those that have metastasized, or as neoadjuvant therapy in certain patients with early-stage ERBB2-positive cases. FDA approval of this drug was based on the findings from the Clinical Evaluation of Pertuzumab and Trastuzumab (CLEOPATRA) trial [38, 43, 46, 51].
Ado–trastuzumab emtansine is a monoclonal antibody linked to an anticancer drug that is used for the treatment of ERBB2 (HER2)-positive recurrent or metastatic breast cancer cases [39, 40].
Lapatinib is a tyrosine kinase inhibitor that blocks the effects of the ERBB2 (HER2) protein and may be used with other drugs to treat ERBB2-positive breast cancers that have progressed after treatment with trastuzumab [52].
The benefit of a combination of two anti-ERBB2 (HER2)-targeted therapies with chemotherapy in early-stage breast cancer is currently being tested in the ongoing phase III ALTTO (Adjuvant Lapatinib and/or Trastuzumab Treatment Optimization) trial, as well as the Neo ALTTO (Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization) [47, 53].
HER2 Testing by RT-PCR
Reverse transcription-polymerase chain reaction (RT-PCR) can be employed to detect ERBB2 gene amplification in breast cancer. Few reports have suggested that RT-PCR testing can be used to confirm FISH test results or to resolve discrepancies between ISH/FISH and IHC (i.e., ISH/FISH negative, IHC 3+). Several studies have shown a concordance of up to 94 % between FISH and RT-PCR results while other authors have reported a high number of false-negative results for ERBB2 with RT-PCR [54, 55]. To date there is insufficient evidence to support qRT-PCR as a substitute for the traditional methods (IHC/FISH) for ERBB2 testing with a possible limitation related to quality assurance. For the small group of patients with equivocal results by both techniques (IHC and FISH), research into novel testing methods (such as qRT-PCR) might be of value [48].
Molecular Classification of Breast Cancers
The most recent advances in molecular pathology based on microarray gene expression studies showed that there is a heterogeneous group of breast cancers with distinct molecular features [49, 50, 56]. In their seminal research work, Perou, Sorlie, and colleagues proposed the first innovative molecular classification of breast cancer based on gene cluster subsets [16, 44]. The newly described “intrinsic” gene cluster subsets included genes of epithelial origin (ESR1, GATA–binding protein–3, X–box–binding protein–1, and hepatocyte nuclear factor–3a), ERBB2, basal epithelial cell markers (cytokeratin (CK) 5 and 17, integrin-b4, and laminin), and “normal breast” markers [57]. Initially four breast cancer “intrinsic” subtypes (basal-like, ERBB2 (HER2)-enriched, luminal, and normal breast-like) were described. Luminal breast cancers were subsequently subclassified into luminal A and luminal B and later studies identified additional “intrinsic” molecular subtypes, such as interferon-rich, claudin-low, and molecular apocrine. The major four molecular subtypes of breast cancers are presented in Table 9.6 [49, 50, 58–60].
Table 9.6
Molecular “intrinsic” subtypes of breast carcinoma (adapted after Cornejo KM, Kandil D, Khan A, Cosar EF. Theranostic and molecular classification of breast cancer. Arch Pathol Lab Med. 2014 Jan;138(1):44–56 and Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013 Sep; 24(9):2206–23)
Molecular subtype | Luminal A | Luminal B | ERBB2 (HER2) enriched | Basal-like |
Frequency (%) | 50–60 % | 10–20 % | 15–20 % | 10–20 % |
Histologic grade | Low grade | High grade | High grade | High grade |
+/−apocrine features | ||||
IHC markers | ESR1—positive | ERBB2 (HER2)—negative | ESR1—negative | ESR1—negative |
PGR—negative | ESR1—positive | PGR—negative | PGR—negative | |
ERBB2 (HER2)—negative | PGR—negative/low | ERBB2 (HER2)—positive | ERBB2 (HER2)—negative | |
Ki67—low | Ki67—high | Ki67—high | Ki67—high | |
Basal markers—negative | Basal markers—negative | Basal markers—negative/positive | Basal markers—positive | |
ERBB2 (HER2)—positive | ||||
ESR1—positive | ||||
PGR—positive/negative | ||||
Ki67—any | ||||
Basal markers—negative | ||||
Genetic expression profile | High ESR1 gene expression | Low ESR1 gene expression | ERBB2 (HER2)-related genes | Basal signature |
Low proliferation gene expression | High proliferation gene expression | High proliferation gene expression | High proliferation gene expression | |
CK | ||||
P-cadherin | ||||
CAV 1/2 | ||||
CD44 | ||||
KIT (CKIT) | ||||
Prognosis | Good | Intermediate or poor | Poor | Poor |
Benefit from chemotherapy | Low (0–5 % pathologic complete response (pCR)) | Intermediate (10–20 % pCR) | Intermediate (25–40 % pCR) | High (≥40 % pCR) |
Treatment | Endocrine therapy for all patients +/− cytotoxics | ERBB2 (HER2) negative: Endocrine therapy for all patients, cytotoxic therapy for most | Cytotoxics + anti-ERBB2 (HER2) | Cytotoxics |
ERBB2 (HER2) positive: Cytotoxics + anti-ERBB2/HER2+ endocrine therapy |
The “breast–like” subgroup of tumors express non-epithelial cell genes, such as adipose tissue-related genes, that cluster with healthy breast and fibroadenomas. The clinical significance of this group needs further elucidation, some researchers favoring this subtype to be a false category related to misinterpretation of poor tissue sampling [16, 59, 61, 62]. Tumors of the “claudin–low” subtype show low expression of cell-cell junction proteins (claudin-3, 4, 7 and E-cadherin) and overexpression of genes involved in immune responses and linked to mesenchymal differentiation. The majority of these tumors are ESR1, PGR, and ERBB2 (HER2) negative with up to 20 % of the cases positive for hormone receptors. Claudin-low tumors are histologically of high grade and have a poor prognosis [6]. The “molecular apocrine” subtype shows similarities to the ERBB2-enhanced subtype with the addition of androgen receptor (AR) activation while the “interferon–rich” subtype is characterized by STAT1 activation [15].
Based on additional molecular studies (genomic DNA copy number arrays, DNA methylation, exon sequencing, mRNA arrays, microRNA sequencing, and reverse-phase protein arrays) the Cancer Genome Atlas Network reported even more new molecular findings in breast cancer. Up to 10 % of all breast cancers were found to have somatic mutations involving three main genes (TP53, PIK3CA, and GATA3). Specific mutations involving GATA3, PIK3CA, and MAP3K1 were described in the luminal A molecular subtype. HER2-positive tumors were further divided into two ERBB2 (HER2) protein expression-defined subgroups with specific signaling pathways. For the basal-like subtype many molecular similarities with high-grade serous ovarian tumors were reported, suggesting a related etiology and the possibility of similar therapeutic approaches [63]. Other novel mutations reported in breast cancer included those in TBX3, RUNX1, CBFB, AFF2, PIK3R1, PTPN22, PTPRD, NF1, SF3B1, and CCND3. Of note, several overlaps with mutations previously described in other diseases were seen. For example, RUNX1 and CBFB showed the same mutations as previously described in acute myeloid leukemia. PIK3R1 mutations were similar to those previously described in glioma and endometrial cancer. The same SF3B1 mutations previously reported in myelodysplastic syndromes and chronic leukocytic leukemia were identified [63].
Other Tumor Markers
TP53
TP53 is a tumor-suppressor gene that is mutated in approximately 30 % of sporadic breast cancer cases. P53 regulates the cellular response to damage and activates genes that are involved in apoptosis, cell cycle arrest, and DNA repair. Mutations of TP53 gene are seen in invasive and in situ breast carcinomas, with a large majority of “basal” phenotype. Recent studies showed that TP53 gene mutations were predicting a worse prognosis and poor response to systemic therapy and that the presence of TP53 mutations/deletions was particularly prognostic in node-negative, ESR1-positive patients. TP53 mutation status correlates with P53 nuclear expression by IHC [15, 44, 64]. Per 2007 ASCO/CAP guidelines for testing in breast cancer there is insufficient data to suggest the use of TP53 for management of breast cancer patients [31].
Ki-67 (MKI67)
Ki-67 is a nuclear protein that correlates with tumor cell proliferation and can be quantified by IHC with the MIB-1 antibody. Although the association of increased proliferation and poor prognosis is well documented, the prognostic role of Ki-67 was evaluated in many studies and showed a modest utility [65–67]. It has been shown that increased Ki-67 expression is associated with ESR1 negativity in breast tumors and is currently used to distinguish luminal A from luminal B molecular subtypes. Treatment-induced alterations of Ki-67 correlate with disease-free and overall survival of breast cancer patients [68, 69]. Thus, tumors with higher expression of Ki-67 and poorer prognosis appear to benefit more from treatment with letrozole than with tamoxifen [70]. Despite the continuous efforts to improve interlaboratory reproducibility of Ki-67 interpretation by IHC, no cutoff point for a positive result has yet been established [16, 65, 71]. Due to lack of standardization in scoring ASCO/CAP do not recommend the use of Ki-67 in clinical practice [72].
EGFR
EGFR (Her1, ERBB1, EGFR) is a cell-surface receptor for the epidermal growth factor group of extracellular protein ligands. Overexpression of EGFR is seen in up to 10 % of invasive breast cancers and is usually reported in ESR1– and PGR-negative tumors and most commonly basal-like molecular subtypes. The predictive and prognostic value of EGFR is still under investigation. The EGFR mutations involving exons 19 and 21 seen in lung carcinomas have not been identified in breast carcinomas and benefit from cetuximab therapy remains to be established [16]. Until further studies, EGFR has a limited, diagnostic-only role.
Cyclin D1 and Cyclin E (CCND1 and CCNE1)
Approximately 50 % of breast cancers are associated with cyclin D1 overexpression and only a minority of cases with CCND1 gene amplification [65, 73]. Its prognostic utility remains controversial and needs to be further investigated. Overexpression of cyclin D1 was reported to be associated with a favorable prognosis [74] while gene amplification with poor outcome [75] and both with a poorer response to endocrine therapy [76]. Cyclin E overexpression appears to be inversely related with ESR1 expression, and associated with poor prognosis, less benefit from endocrine therapy, and increased sensitivity to chemotherapy.
uPA and PAI-1 (PLAU and SERPINE1)
Recent studies showed that the urokinase plasminogen activator (uPA)/plasminogen activator inhibitor (PAI-1) is involved in breast tumor invasion, angiogenesis, and metastasis. High uPA/PAI-I levels were reported in ESR1-negative tumors of high histologic grade. Low levels of expression have been associated with low recurrence risk and minimal benefit from adjuvant chemotherapy. Both markers emerged as independent predictors of tumor relapse and poor overall survival in breast cancers independent of lymph node status [65]. ELISA testing is the best available technique to determine uPA/PAI-1 and the results correlate well with the clinical outcome. Fresh or frozen tumor tissue samples are needed for testing, limiting its use in clinical practice [31, 77].
FOXA1
In breast cancer FOXA1 expression may play a role in identifying ESR1-positive tumors that undergo rapid reprogramming of the estrogen receptor signaling pathway. In these tumors FOXA1 expression is associated with poor outcome and resistance to therapy. In contrast, in ESR1-negative breast tumors FOXA1 is highly correlated with low-grade histology and disease-free survival. Although still limited supportive data is available, it was suggested that FOXA1 may play a future role as an immunohistochemical and/or clinical marker for luminal A subtype breast cancers [31].
Topoisomerase IIα
Topoisomerase IIα (TOP2A) overexpression is reported in luminal B, ERBB2 (HER2)-enriched, and basal-like breast tumors. Anthracyclines inhibit TOP2A activity and several studies have evaluated the predictive value of TOP2A with inconsistent results. Both overexpression and deletion of TOP2A were seen in breast tumors showing a favorable response to chemotherapeutics [78, 79]. TOP2A FISH pharmDxTM Kit is an FDA-approved FISH-based test designed to detect amplifications and deletions of the TOP2A gene on formalin-fixed, paraffin-embedded tumor samples. Both deletions and amplifications of the TOP2A gene serve as markers of poor prognosis in high-risk breast cancer patients. The results from this test can be used as an adjunct to the clinicopathologic data [78, 79].
Circulating and Disseminated Tumor Cells
Studies have shown that the number of circulating tumor cell (CTC) is associated with lymph node involvement and disease-free survival in early breast cancer and with progression-free and overall survival in metastatic disease. These findings can have implications for assays targeting epithelial antigens that may miss part of the circulating cell populations. The CellSearch CTC detection system (Veridex, LLC) is FDA approved for use in metastatic breast cancer. The test uses an immunomagnetic system to isolate epithelial cells from the peripheral blood and offers a count of CTCs per 7.5 mL blood sample. Greater than five CTCs per 7.5 mL sample have been associated with worse progression-free and overall survivals. SWOG S0500 clinical trial will further determine if the CTC level can be used to assess the benefits from chemotherapy in metastatic breast cancer cases. Until those results are available, this test is not recommended by the ASCO breast cancer tumor marker panel and is not included in the breast cancer NCCN guidelines [80].
Multigene Expression Assays
A variety of multigene and multiprotein expression assays are available for the identification of tumor subtypes and for the assessment of prognosis and likelihood of response to specific treatments. The majority are proprietary assays developed and performed by a single laboratory. Multigene assays can detect gene expression patterns by several techniques including qRT-PCR; hybridization of labeled tumoral nucleic acids to small, immobilized, synthetic DNA strands (microarrays); FISH; and immunohistochemistry [32, 81, 82]. The aim of gene expression profiling is to further assist clinician in objectively estimating the outcome by providing a better prediction than the traditional clinical and pathologic parameters. Pathologists have the option to include the results of these assays in their reports [83]. Several gene signatures with prognostic and predictive value in breast cancer are available for use and algorithms for inclusion of these assays in clinical therapy decision making have been created [16].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree